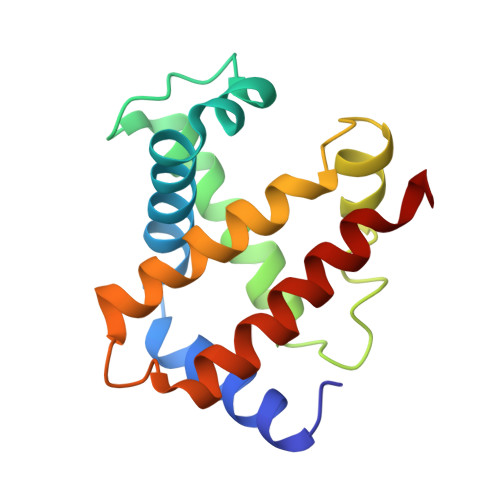
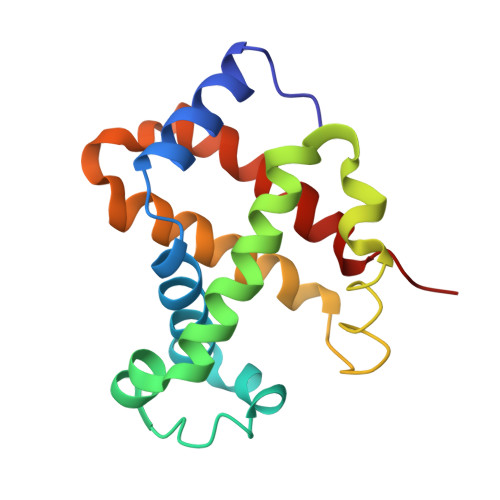
Crystal structure of hemoglobin from mouse (Mus musculus) compared with those from other small animals and humans.
Sundaresan, S.S., Ramesh, P., Shobana, N., Vinuchakkaravarthy, T., Yasien, S., Ponnuswamy, M.N.G.(2021) Acta Crystallogr F Struct Biol Commun 77: 113-120
- PubMed: 33830076
- DOI: https://doi.org/10.1107/S2053230X2100306X
- Primary Citation of Related Structures:
3HRW - PubMed Abstract:
Mice (Mus musculus) are nocturnal small animals belonging to the rodent family that live in burrows, an environment in which significantly high CO 2 levels prevail. It is expected that mouse hemoglobin (Hb) plays an important role in their adaptation to living in such a high-CO 2 environment, while many other species cannot. In the present study, mouse Hb was purified and crystallized at a physiological pH of 7 in the orthorhombic space group P2 1 2 1 2 1 ; the crystals diffracted to 2.8 Å resolution. The primary amino-acid sequence and crystal structure of mouse Hb were compared with those of mammalian Hbs in order to investigate the structure-function relationship of mouse Hb. Differences were observed from guinea pig Hb in terms of amino-acid sequence and from cat Hb in overall structure (in terms of r.m.s.d.). The difference in r.m.s.d. from cat Hb may be due to the existence of the molecule in a conformation other than the R-state. Analysis of tertiary- and quaternary-structural features, the α1β2 interface region and the heme environment without any ligands in all four heme groups showed that mouse methemoglobin is in an intermediate state between the R-state and the T-state that is much closer to the R-state conformation.
Organizational Affiliation:
Department of Physics, Sir Theagaraya College, Chennai 600 021, India.