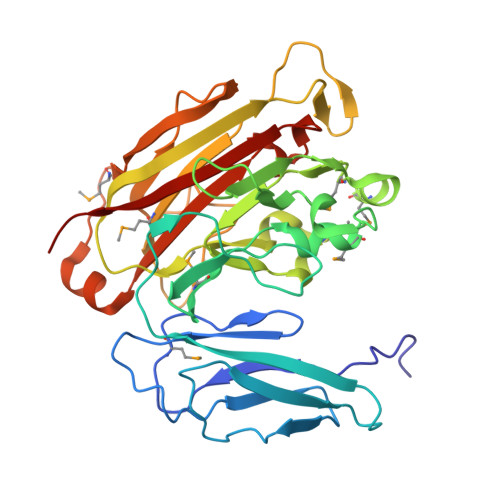
Structure of Bacteroides thetaiotaomicron BT2081 at 2.05 A resolution: the first structural representative of a new protein family that may play a role in carbohydrate metabolism.
Yeh, A.P., Abdubek, P., Astakhova, T., Axelrod, H.L., Bakolitsa, C., Cai, X., Carlton, D., Chen, C., Chiu, H.J., Chiu, M., Clayton, T., Das, D., Deller, M.C., Duan, L., Ellrott, K., Farr, C.L., Feuerhelm, J., Grant, J.C., Grzechnik, A., Han, G.W., Jaroszewski, L., Jin, K.K., Klock, H.E., Knuth, M.W., Kozbial, P., Krishna, S.S., Kumar, A., Lam, W.W., Marciano, D., McMullan, D., Miller, M.D., Morse, A.T., Nigoghossian, E., Nopakun, A., Okach, L., Puckett, C., Reyes, R., Tien, H.J., Trame, C.B., van den Bedem, H., Weekes, D., Wooten, T., Xu, Q., Hodgson, K.O., Wooley, J., Elsliger, M.A., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1287-1296
- PubMed: 20944224
- DOI: https://doi.org/10.1107/S1744309110028228
- Primary Citation of Related Structures:
3HBZ - PubMed Abstract:
BT2081 from Bacteroides thetaiotaomicron (GenBank accession code NP_810994.1) is a member of a novel protein family consisting of over 160 members, most of which are found in the different classes of Bacteroidetes. Genome-context analysis lends support to the involvement of this family in carbohydrate metabolism, which plays a key role in B. thetaiotaomicron as a predominant bacterial symbiont in the human distal gut microbiome. The crystal structure of BT2081 at 2.05 Å resolution represents the first structure from this new protein family. BT2081 consists of an N-terminal domain, which adopts a β-sandwich immunoglobulin-like fold, and a larger C-terminal domain with a β-sandwich jelly-roll fold. Structural analyses reveal that both domains are similar to those found in various carbohydrate-active enzymes. The C-terminal β-jelly-roll domain contains a potential carbohydrate-binding site that is highly conserved among BT2081 homologs and is situated in the same location as the carbohydrate-binding sites that are found in structurally similar glycoside hydrolases (GHs). However, in BT2081 this site is partially occluded by surrounding loops, which results in a deep solvent-accessible pocket rather than a shallower solvent-exposed cleft.
Organizational Affiliation:
Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, Menlo Park,CA, USA.