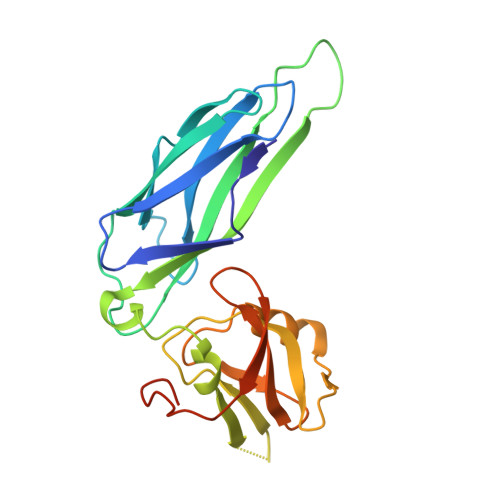
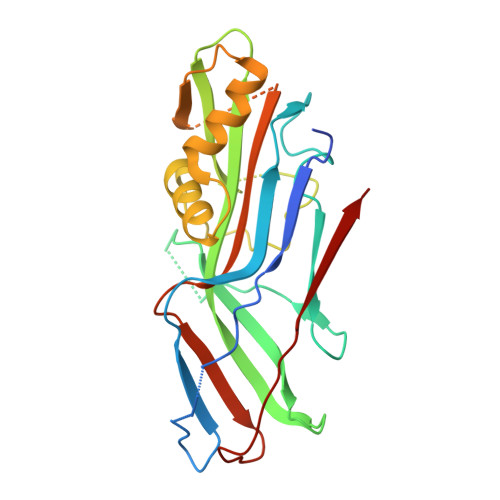
Structural and thermodynamic characterization of pre- and postpolymerization states in the F4 fimbrial subunit FaeG
Van Molle, I., Moonens, K., Garcia-Pino, A., Buts, L., De Kerpel, M., Wyns, L., Bouckaert, J., De Greve, H.(2009) J Mol Biol 394: 957-967
- PubMed: 19799915
- DOI: https://doi.org/10.1016/j.jmb.2009.09.059
- Primary Citation of Related Structures:
3GEA, 3GEW, 3GFU, 3GGH, 3HLR - PubMed Abstract:
Enterotoxigenic Escherichia coli expressing F4 fimbriae are the major cause of porcine colibacillosis and are responsible for significant death and morbidity in neonatal and postweaned piglets. Via the chaperone-usher pathway, F4 fimbriae are assembled into thin, flexible polymers mainly composed of the single-domain adhesin FaeG. The F4 fimbrial system has been labeled eccentric because the F4 pilins show some features distinct from the features of pilins of other chaperone-usher-assembled structures. In particular, FaeG is much larger than other pilins (27 versus approximately 17 kDa), grafting an additional carbohydrate binding domain on the common immunoglobulin-like core. Structural data of FaeG during different stages of the F4 fimbrial biogenesis process, combined with differential scanning calorimetry measurements, confirm the general principles of the donor strand complementation/exchange mechanisms taking place during pilus biogenesis via the chaperone-usher pathway.
Organizational Affiliation:
Structural Biology Brussels, Vrije Universiteit Brussel, Pleinlaan 2, 1050 Brussels, Belgium.