Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif
Klenchin, V.A., Czyz, A., Goryshin, I.Y., Gradman, R., Lovell, S., Rayment, I., Reznikoff, W.S.(2008) Nucleic Acids Res 36: 5855-5862
- PubMed: 18790806
- DOI: https://doi.org/10.1093/nar/gkn577
- Primary Citation of Related Structures:
3ECP - PubMed Abstract:
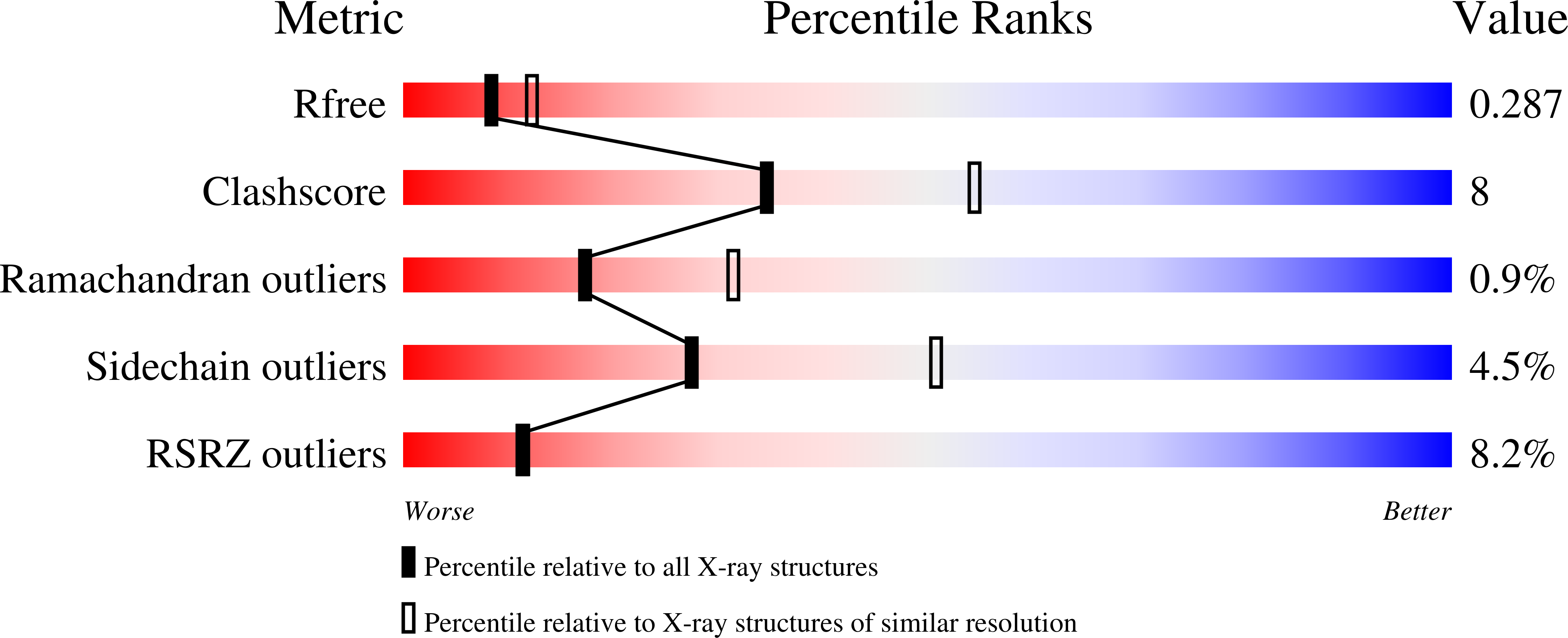
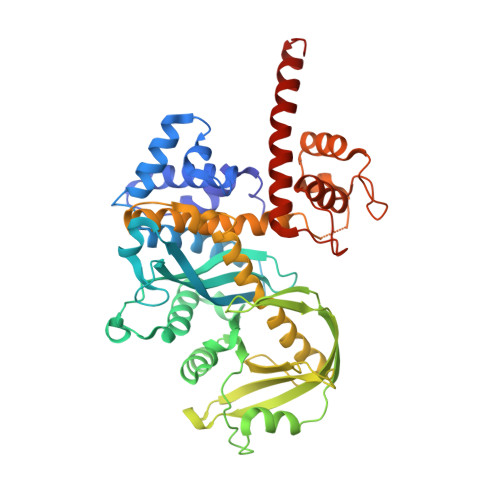
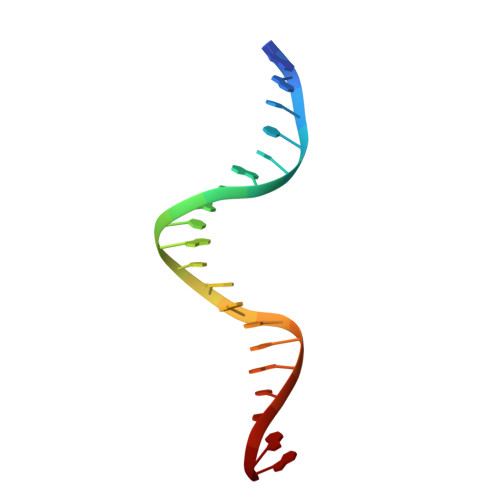
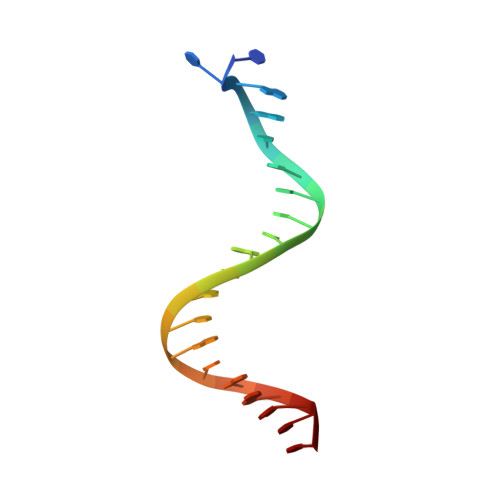
Bacterial DNA transposition is an important model system for studying DNA recombination events such as HIV-1 DNA integration and RAG-1-mediated V(D)J recombination. This communication focuses on the role of protein-phosphate contacts in manipulating DNA structure as a requirement for transposition catalysis. In particular, the participation of the nontransferred strand (NTS) 5' phosphate in Tn5 transposition strand transfer is analyzed. The 5' phosphate plays no direct catalytic role, nonetheless its presence stimulates strand transfer approximately 30-fold. X-ray crystallography indicates that transposase-DNA complexes formed with NTS 5' phosphorylated DNA have two properties that contrast with structures formed with complexes lacking the 5' phosphate or complexes generated from in-crystal hairpin cleavage. Transposase residues R210, Y319 and R322 of the (R)YREK motif coordinate the 5' phosphate rather than the subterminal NTS phosphate, and the 5' NTS end is moved away from the 3' transferred strand end. Mutation R210A impairs the 5' phosphate stimulation. It is posited that DNA phosphate coordination by R210, Y319 and R322 results in movement of the 5' NTS DNA away from the 3'-end thus allowing efficient target DNA binding. It is likely that this role for the newly identified RYR triad is utilized by other transposase-related proteins.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin at Madison, 433 Babcock Drive, Madison, WI 53706, USA.