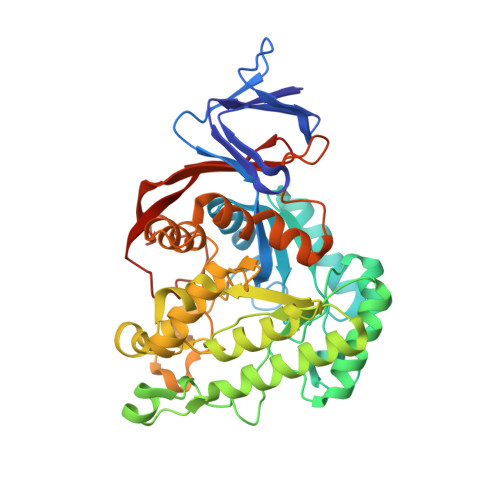
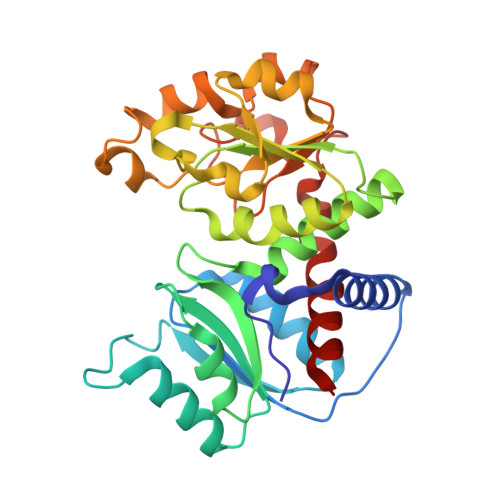
Dihydroorotase from the hyperthermophile Aquifiex aeolicus is activated by stoichiometric association with aspartate transcarbamoylase and forms a one-pot reactor for pyrimidine biosynthesis.
Zhang, P., Martin, P.D., Purcarea, C., Vaishnav, A., Brunzelle, J.S., Fernando, R., Guy-Evans, H.I., Evans, D.R., Edwards, B.F.(2009) Biochemistry 48: 766-778
- PubMed: 19128030
- DOI: https://doi.org/10.1021/bi801831r
- Primary Citation of Related Structures:
3D6N - PubMed Abstract:
In prokaryotes, the first three enzymes in pyrimidine biosynthesis, carbamoyl phosphate synthetase (CPS), aspartate transcarbamoylase (ATC), and dihydroorotase (DHO), are commonly expressed separately and either function independently (Escherichia coli) or associate into multifunctional complexes (Aquifex aeolicus). In mammals the enzymes are expressed as a single polypeptide chain (CAD) in the order CPS-DHO-ATC and associate into a hexamer. This study presents the three-dimensional structure of the noncovalent hexamer of DHO and ATC from the hyperthermophile A. aeolicus at 2.3 A resolution. It is the first structure of any multienzyme complex in pyrimidine biosynthesis and is a possible model for the core of mammalian CAD. The structure has citrate, a near isosteric analogue of carbamoyl aspartate, bound to the active sites of both enzymes. Three active site loops that are intrinsically disordered in the free, inactive DHO are ordered in the complex. The reorganization also changes the peptide bond between Asp153, a ligand of the single zinc atom in DHO, and Gly154, to the rare cis conformation. In the crystal structure, six DHO and six ATC chains form a hollow dodecamer, in which the 12 active sites face an internal reaction chamber that is approximately 60 A in diameter and connected to the cytosol by narrow tunnels. The entrances and the interior of the chamber are both electropositive, which suggests that the architecture of this nanoreactor modifies the kinetics of the bisynthase, not only by steric channeling but also by preferential escape of the product, dihydroorotase, which is less negatively charged than its precursors, carbamoyl phosphate, aspartate, or carbamoyl aspartate.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Wayne State University School of Medicine, 540 East Canfield Street, Detroit, Michigan 48201, USA.