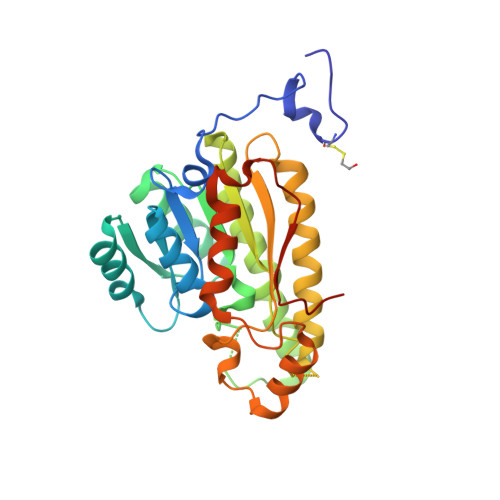
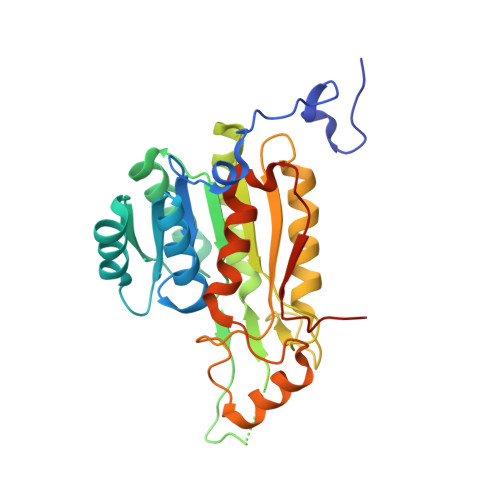
Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Zhang, R., Zhu, G., Zhang, W., Cao, S., Ou, X., Li, X., Bartlam, M., Xu, Y., Zhang, X.C., Rao, Z.(2008) Protein Sci 17: 1412-1423
- PubMed: 18566346
- DOI: https://doi.org/10.1110/ps.035089.108
- Primary Citation of Related Structures:
3CTM - PubMed Abstract:
A novel short-chain (S)-1-phenyl-1,2-ethanediol dehydrogenase (SCR) from Candida parapsilosis exhibits coenzyme specificity for NADPH over NADH. It catalyzes an anti-Prelog type reaction to reduce 2-hydroxyacetophenone into (S)-1-phenyl-1,2-ethanediol. The coding gene was overexpressed in Escherichia coli and the purified protein was crystallized. The crystal structure of the apo-form was solved to 2.7 A resolution. This protein forms a homo-tetramer with a broken 2-2-2 symmetry. The overall fold of each SCR subunit is similar to that of the known structures of other homologous alcohol dehydrogenases, although the latter usually form tetramers with perfect 2-2-2 symmetries. Additionally, in the apo-SCR structure, the entrance of the NADPH pocket is blocked by a surface loop. In order to understand the structure-function relationship of SCR, we carried out a number of mutagenesis-enzymatic analyses based on the new structural information. First, mutations of the putative catalytic Ser-Tyr-Lys triad confirmed their functional role. Second, truncation of an N-terminal 31-residue peptide indicated its role in oligomerization, but not in catalytic activity. Similarly, a V270D point mutation rendered the SCR as a dimer, rather than a tetramer, without affecting the enzymatic activity. Moreover, the S67D/H68D double-point mutation inside the coenzyme-binding pocket resulted in a nearly 10-fold increase and a 20-fold decrease in the k(cat) /K(M) value when NADH and NADPH were used as cofactors, respectively, with k(cat) remaining essentially the same. This latter result provides a new example of a protein engineering approach to modify the coenzyme specificity in SCR and short-chain dehydrogenases/reductases in general.
Organizational Affiliation:
Key Laboratory of Industrial Biotechnology of Ministry of Education and School of Biotechnology, Jiangnan University, Wuxi 214122, People's Republic of China.