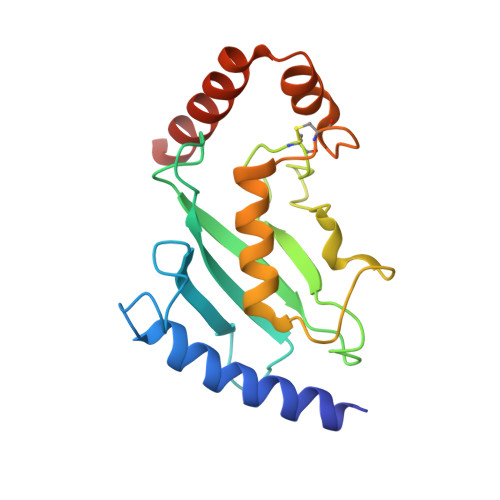
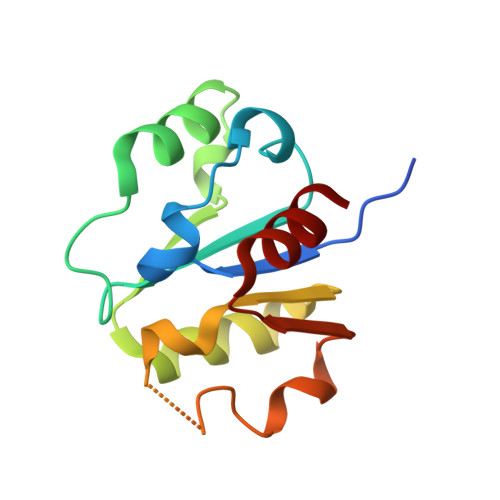
Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Williams, C., van den Berg, M., Panjikar, S., Stanley, W.A., Distel, B., Wilmanns, M.(2011) EMBO J 31: 391
- PubMed: 22085930
- DOI: https://doi.org/10.1038/emboj.2011.411
- Primary Citation of Related Structures:
2Y9M - PubMed Abstract:
Ubiquitin-conjugating enzymes (E2s) coordinate distinct types of ubiquitination via specific E3 ligases, to a large number of protein substrates. While many E2 enzymes need only the presence of an E3 ligase for substrate ubiquitination, a number of E2s require additional, non-canonical binding partners to specify their function. Here, we have determined the crystal structure and function of an E2/co-activator assembly, the Pex4p:Pex22p complex. The peroxisome-associated E2 enzyme Pex4p binds the peroxisomal membrane protein Pex22p through a binding site that does not overlap with any other known interaction interface in E2 enzymes. Pex22p association enhances Pex4p's ability to transfer ubiquitin to a substrate in vitro, and Pex22p binding-deficient forms of Pex4p are unable to ubiquitinate the peroxisomal import receptor Pex5p in vivo. Our data demonstrate that the Pex4p:Pex22p assembly, and not Pex4p alone, functions as the E2 enzyme required for Pex5p ubiquitination, establishing a novel mechanism of E2 enzyme regulation.
Organizational Affiliation:
Structural Biology Unit, European Molecular Biology Laboratory, Hamburg, Germany. c.williams@embl-hamburg.de