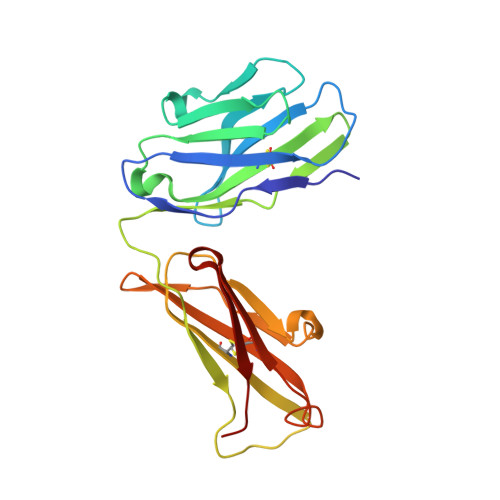
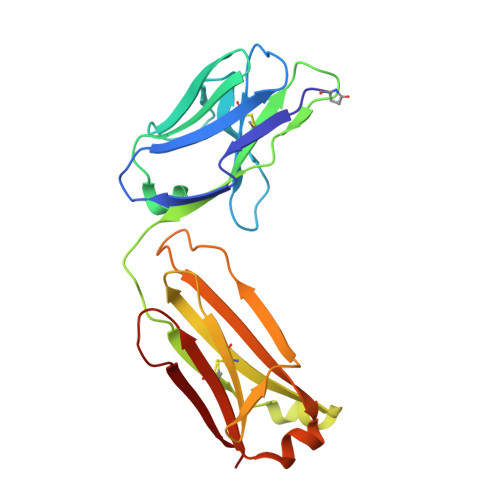
Crystal Structure of Human Prion Protein Bound to a Therapeutic Antibody.
Antonyuk, S.V., Trevitt, C.R., Strange, R.W., Jackson, G.S., Sangar, D., Batchelor, M., Cooper, S., Fraser, C., Jones, S., Georgiou, T., Khalili-Shirazi, A., Clarke, A.R., Hasnain, S.S., Collinge, J.(2009) Proc Natl Acad Sci U S A 106: 2554
- PubMed: 19204296
- DOI: https://doi.org/10.1073/pnas.0809170106
- Primary Citation of Related Structures:
2W9D, 2W9E - PubMed Abstract:
Prion infection is characterized by the conversion of host cellular prion protein (PrP(C)) into disease-related conformers (PrP(Sc)) and can be arrested in vivo by passive immunization with anti-PrP monoclonal antibodies. Here, we show that the ability of an antibody to cure prion-infected cells correlates with its binding affinity for PrP(C) rather than PrP(Sc). We have visualized this interaction at the molecular level by determining the crystal structure of human PrP bound to the Fab fragment of monoclonal antibody ICSM 18, which has the highest affinity for PrP(C) and the highest therapeutic potency in vitro and in vivo. In this crystal structure, human PrP is observed in its native PrP(C) conformation. Interactions between neighboring PrP molecules in the crystal structure are mediated by close homotypic contacts between residues at position 129 that lead to the formation of a 4-strand intermolecular beta-sheet. The importance of this residue in mediating protein-protein contact could explain the genetic susceptibility and prion strain selection determined by polymorphic residue 129 in human prion disease, one of the strongest common susceptibility polymorphisms known in any human disease.
Organizational Affiliation:
Molecular Biophysics Group, Science and Technology Facilities Council, Daresbury Laboratory, Warrington, Cheshire WA4 4AD, United Kingdom.