Elucidating the role of disulfide bond on amyloid formation and fibril reversibility of somatostatin-14: relevance to its storage and secretion.
Anoop, A., Ranganathan, S., Das Dhaked, B., Jha, N.N., Pratihar, S., Ghosh, S., Sahay, S., Kumar, S., Das, S., Kombrabail, M., Agarwal, K., Jacob, R.S., Singru, P., Bhaumik, P., Padinhateeri, R., Kumar, A., Maji, S.K.(2014) J Biol Chem 289: 16884-16903
- PubMed: 24782311
- DOI: https://doi.org/10.1074/jbc.M114.548354
- Primary Citation of Related Structures:
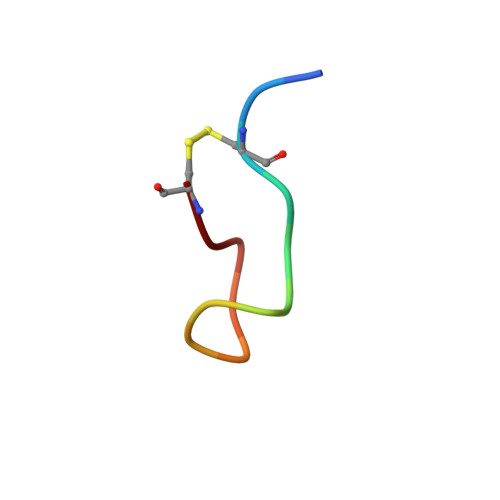
2MI1 - PubMed Abstract:
The storage of protein/peptide hormones within subcellular compartments and subsequent release are crucial for their native function, and hence these processes are intricately regulated in mammalian systems. Several peptide hormones were recently suggested to be stored as amyloids within endocrine secretory granules. This leads to an apparent paradox where storage requires formation of aggregates, and their function requires a supply of non-aggregated peptides on demand. The precise mechanism behind amyloid formation by these hormones and their subsequent release remain an open question. To address this, we examined aggregation and fibril reversibility of a cyclic peptide hormone somatostatin (SST)-14 using various techniques. After proving that SST gets stored as amyloid in vivo, we investigated the role of native structure in modulating its conformational dynamics and self-association by disrupting the disulfide bridge (Cys(3)-Cys(14)) in SST. Using two-dimensional NMR, we resolved the initial structure of somatostatin-14 leading to aggregation and further probed its conformational dynamics in silico. The perturbation in native structure (S-S cleavage) led to a significant increase in conformational flexibility and resulted in rapid amyloid formation. The fibrils formed by disulfide-reduced noncyclic SST possess greater resistance to denaturing conditions with decreased monomer releasing potency. MD simulations reveal marked differences in the intermolecular interactions in SST and noncyclic SST providing plausible explanation for differential aggregation and fibril reversibility observed experimentally in these structural variants. Our findings thus emphasize that subtle changes in the native structure of peptide hormone(s) could alter its conformational dynamics and amyloid formation, which might have significant implications on their reversible storage and secretion.
Organizational Affiliation:
From the Department of Biosciences and Bioengineering, Indian Institute of Technology Bombay, Mumbai 400 076.