Solution structure of HP1264, Complex I subunit E from Helicobacter pylori, reveals a novel fold for FMN binding protein
Lee, K.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NADH dehydrogenase I subunit E | 76 | Helicobacter pylori 26695 | Mutation(s): 0 Gene Names: HP_1264 | 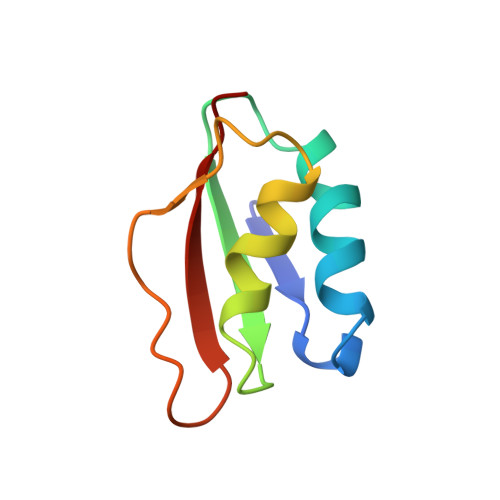 | |
UniProt | |||||
Find proteins for O25854 (Helicobacter pylori (strain ATCC 700392 / 26695)) Explore O25854 Go to UniProtKB: O25854 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O25854 | ||||
Sequence AnnotationsExpand | |||||
| |||||