Solution structure and dynamics of ERp18, a small endoplasmic reticulum resident oxidoreductase .
Rowe, M.L., Ruddock, L.W., Kelly, G., Schmidt, J.M., Williamson, R.A., Howard, M.J.(2009) Biochemistry 48: 4596-4606
- PubMed: 19361226
- DOI: https://doi.org/10.1021/bi9003342
- Primary Citation of Related Structures:
2K8V - PubMed Abstract:
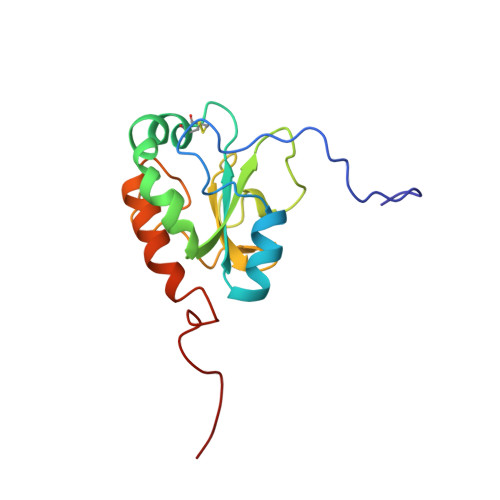
Here we report the solution structure of oxidized ERp18 as determined using NMR spectroscopy. ERp18 is the smallest member of the protein disulfide isomerase (PDI) family of proteins to contain a Cys-Xxx-Xxx-Cys active site motif. It is an 18 kDa endoplasmic reticulum resident protein with unknown function although sequence similarity to individual domains of the thiol-disulfide oxidoreductase PDI suggests ERp18 may have a similar structure and function. Like the catalytic domains of PDI, ERp18 adopts a thioredoxin fold with a thioredoxin-like active site located at the N-terminus of a long kinked helix that spans the length of the protein. Comparison of backbone chemical shifts for oxidized and reduced ERp18 shows the majority of residues possess the same backbone conformation in both states, with differences limited to the active site and regions in close proximity. S(2) order parameters from NMR backbone dynamics were found to be 0.81 for oxidized and 0.91 for reduced ERp18, and these observations, in combination with amide hydrogen exchange rates, imply a more rigid and compact backbone for the reduced structure. These observations support a putative role for ERp18 within the cell as an oxidase, introducing disulfide bonds to substrate proteins, providing structural confirmation of ERp18's role as a thiol-disulfide oxidoreductase.
Organizational Affiliation:
Department of Biosciences, University of Kent, Canterbury, Kent CT2 7NJ, United Kingdom.