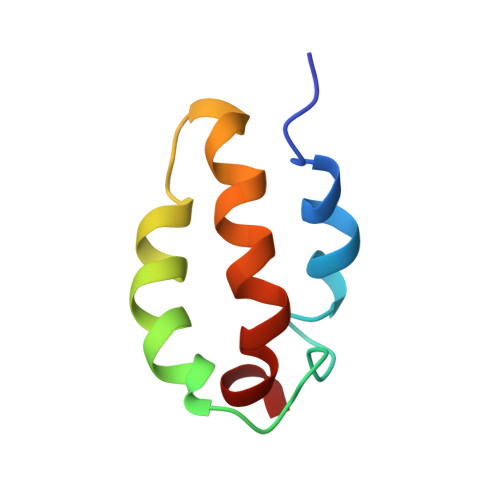
NMR Solution Conformations and Interactions of Integrin {alpha}L{beta}2 Cytoplasmic Tails
Bhunia, A., Tang, X.-Y., Mohanram, H., Tan, S.-M., Bhattacharjya, S.(2009) J Biol Chem 284: 3873-3884
- PubMed: 19073598
- DOI: https://doi.org/10.1074/jbc.M807236200
- Primary Citation of Related Structures:
2K8O - PubMed Abstract:
The integrins are bi-directional signal transducers. Devoid of enzymatic activity, the integrin cytoplasmic tail serves as a hub for the recruitment of cytosolic proteins, and many of these are signaling molecules. The leukocyte-restricted integrin alphaLbeta2 is essential for the adhesion, migration, and proliferation of leukocytes. Here we report solution conformations and interactions of the alphaLbeta2 cytoplasmic tails by NMR analyses. The alphaL tail is characterized by three helical segments in the order of helix 1-3 that are connected by two loops with helix 3 having a number of nuclear Overhauser effect contacts with helix 1 and helix 2. The conformation of the beta2 tail is less defined with only a helical segment restricted at its N terminus. Acidic residues from the helix 2-loop-helix 3 motif of alphaL were found to be responsible for its binding to calcium ion. There were detectable interactions between alphaL and beta2 tails, involving helix 1 and helix 3 of the alphaL tail and the N-terminal helix of the beta2 tail. Talin head domain that contains the FERM domain showed binding affinity of Kd approximately 0.5 microm with the beta2 tail. The binding affinity of alphaL and beta2 tails is Kd approximately 2.63 microm. These data are in line with the activating property of talin head domain on alphaLbeta2 by which binding of talin head domain to beta2 tail disrupts the interface of the alphaL and beta2 tails that constrains alphaLbeta2 in a resting state.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551, Singapore.