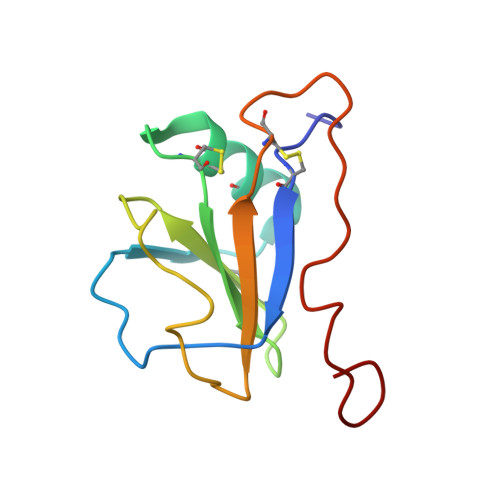
Structure of the leech protein saratin and characterization of its binding to collagen
Gronwald, W., Bomke, J., Maurer, T., Domogalla, B., Huber, F., Schumann, F., Kremer, W., Fink, F., Rysiok, T., Frech, M., Kalbitzer, H.R.(2008) J Mol Biol 381: 913-927
- PubMed: 18585393
- DOI: https://doi.org/10.1016/j.jmb.2008.06.034
- Primary Citation of Related Structures:
2K13 - PubMed Abstract:
The leech protein Saratin from Hirudo medicinalis prevents thrombocyte aggregation by interfering with the first binding step of the thrombocytes to collagen by binding to collagen. We solved the three-dimensional structure of the leech protein Saratin in solution and identified its collagen binding site by NMR titration experiments. The NMR structure of Saratin consists of one alpha-helix and a five-stranded beta-sheet arranged in the topology betabetaalphabetabetabeta. The C-terminal region, of about 20 amino acids in length, adopts no regular structure. NMR titration experiments with collagen peptides show that the collagen interaction of Saratin takes place in a kind of notch that is formed by the end of the alpha-helix and the beta-sheet. NMR data-driven docking experiments to collagen model peptides were used to elucidate the putative binding mode of Saratin and collagen. Mainly, parts of the first and the end of the fifth beta-strand, the loop connecting the alpha-helix and the third beta-strand, and a short part of the loop connecting the fourth and fifth beta-strand participate in binding.
Organizational Affiliation:
Institute of Biophysics and Physical Biochemistry, University of Regensburg, 93040 Regensburg, Germany.