Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target.
Rifat, D., Wright, N.T., Varney, K.M., Weber, D.J., Black, L.W.(2008) J Mol Biol 375: 720-734
- PubMed: 18037438
- DOI: https://doi.org/10.1016/j.jmb.2007.10.064
- Primary Citation of Related Structures:
2JUB - PubMed Abstract:
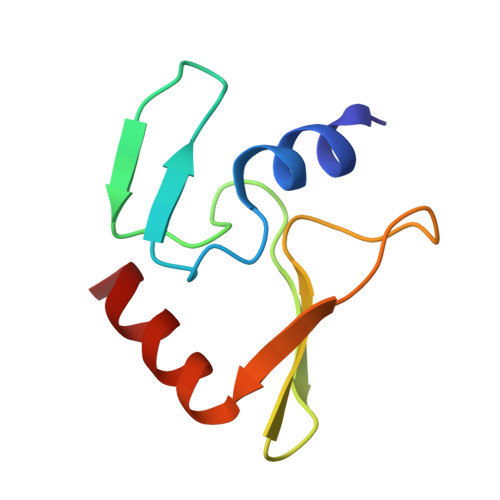
Phage T4 protects its DNA from the two-gene-encoded gmrS/gmrD (glucose-modified hydroxymethylcytosine restriction endonuclease) CT of pathogenic Escherichia coli, CT596, by injecting several hundred copies of the 76-amino-acid-residue nuclease inhibitor, IPI*, into the infected host. Here, the three-dimensional solution structure of mature IPI* is reported as determined by nuclear magnetic resonance techniques using 1290 experimental nuclear Overhauser effect and dipolar coupling constraints ( approximately 17 constraints per residue). Close examination of this oblate-shaped protein structure reveals a novel fold consisting of two small beta-sheets (beta1: B1 and B2; beta2: B3-B5) flanked at the N- and C-termini by alpha-helices (H1 and H2). Such a fold is very compact in shape and allows ejection of IPI* through the narrow 30-A portal and tail tube apertures of the virion without unfolding. Structural and dynamic measurements identify an exposed hydrophobic knob that is a putative gmrS/gmrD-binding site. A single gene from the uropathogenic E. coli UT189, which codes for a gmrS/gmrD-like UT fusion enzyme (with approximately 90% identity to the heterodimeric CT enzyme), has evolved IPI* inhibitor immunity. Analysis of the gmrS/gmrD restriction endonuclease enzyme family and its IPI* family phage antagonists reveals an evolutionary pathway that has elaborated a surprisingly diverse and specifically fitted set of coevolving attack and defense structures.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Maryland School of Medicine, 108 North Greene Street, Baltimore, MD 21201-1503, USA.