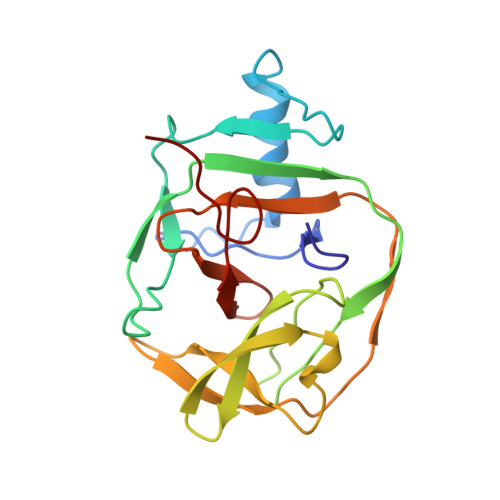
NMR structure of a KlbA intein precursor from Methanococcus jannaschii
Johnson, M.A., Southworth, M.W., Herrmann, T., Brace, L., Perler, F.B., Wuthrich, K.(2007) Protein Sci 16: 1316-1328
- PubMed: 17586768
- DOI: https://doi.org/10.1110/ps.072816707
- Primary Citation of Related Structures:
2JMZ, 2JNQ - PubMed Abstract:
Certain proteins of unicellular organisms are translated as precursor polypeptides containing inteins (intervening proteins), which are domains capable of performing protein splicing. These domains, in conjunction with a single residue following the intein, catalyze their own excision from the surrounding protein (extein) in a multistep reaction involving the cleavage of two intein-extein peptide bonds and the formation of a new peptide bond that ligates the two exteins to yield the mature protein. We report here the solution NMR structure of a 186-residue precursor of the KlbA intein from Methanococcus jannaschii, comprising the intein together with N- and C-extein segments of 7 and 11 residues, respectively. The intein is shown to adopt a single, well-defined globular domain, representing a HINT (Hedgehog/Intein)-type topology. Fourteen beta-strands are arranged in a complex fold that includes four beta-hairpins and an antiparallel beta-ribbon, and there is one alpha-helix, which is packed against the beta-ribbon, and one turn of 3(10)-helix in the loop between the beta-strands 8 and 9. The two extein segments show increased disorder, and form only minimal nonbonding contacts with the intein domain. Structure-based mutation experiments resulted in a proposal for functional roles of individual residues in the intein catalytic mechanism.
Organizational Affiliation:
The Scripps Research Institute, Department of Molecular Biology, La Jolla, CA 92037, USA.