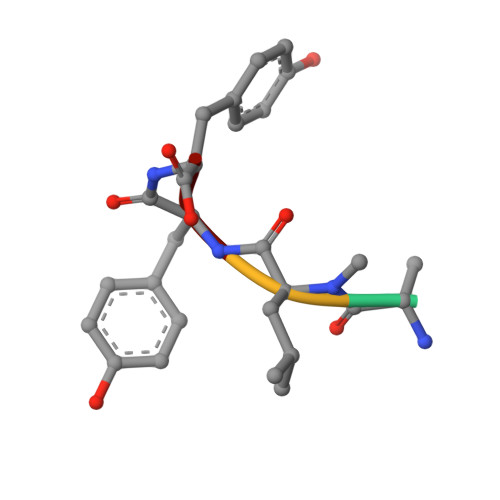
Binding Structure of the Leucine Aminopeptidase Inhibitor Microginin Fr1.
Kraft, M., Schleberger, C., Weckesser, J., Schulz, G.E.(2006) FEBS Lett 580: 6943
- PubMed: 17157838
- DOI: https://doi.org/10.1016/j.febslet.2006.11.060
- Primary Citation of Related Structures:
2J9A - PubMed Abstract:
Natural bioactive compounds are of general interest for pharmaceutical research because they may serve as leads in drug development campaigns. Among them, microginins are linear peptides known to inhibit various exopeptidases. The crystal structure of microginin FR1 from Microcystis sp. bound to bovine lens leucine aminopeptidase was established at 1.73 Angstrom resolution. The observed binding structure could be beneficial for the design of potent aminopeptidase inhibitors.
Organizational Affiliation:
Institut für Biologie II, Microbiologie, Albert-Ludwigs-Universität, Schänzlestr. 1, D-79104 Freiburg im Breisgau, Germany.