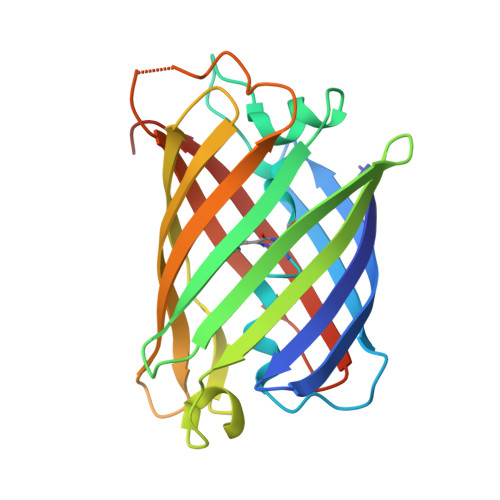
The 2.1A crystal structure of copGFP, a representative member of the copepod clade within the green fluorescent protein superfamily
Wilmann, P.G., Battad, J., Petersen, J., Wilce, M.C.J., Dove, S., Devenish, R.J., Prescott, M., Rossjohn, J.(2006) J Mol Biol 359: 890-900
- PubMed: 16697009
- DOI: https://doi.org/10.1016/j.jmb.2006.04.002
- Primary Citation of Related Structures:
2G3O - PubMed Abstract:
The green fluorescent protein (avGFP), its variants, and the closely related GFP-like proteins are characterized structurally by a cyclic tri-peptide chromophore located centrally within a conserved beta-can fold. Traditionally, these GFP family members have been isolated from the Cnidaria although recently, distantly related GFP-like proteins from the Bilateria, a sister group of the Cnidaria have been described, although no representative structure from this phylum has been reported to date. We have determined to 2.1A resolution the crystal structure of copGFP, a representative GFP-like protein from a copepod, a member of the Bilateria. The structure of copGFP revealed that, despite sharing only 19% sequence identity with GFP, the tri-peptide chromophore (Gly57-Tyr58-Gly59) of copGFP adopted a cis coplanar conformation within the conserved beta-can fold. However, the immediate environment surrounding the chromophore of copGFP was markedly atypical when compared to other members of the GFP-superfamily, with a large network of bulky residues observed to surround the chromophore. Arg87 and Glu222 (GFP numbering 96 and 222), the only two residues conserved between copGFP, GFP and GFP-like proteins are involved in autocatalytic genesis of the chromophore. Accordingly, the copGFP structure provides an alternative platform for the development of a new suite of fluorescent protein tools. Moreover, the structure suggests that the autocatalytic genesis of the chromophore is remarkably tolerant to a high degree of sequence and structural variation within the beta-can fold of the GFP superfamily.
Organizational Affiliation:
The Protein Crystallography Unit, School of Biomedical Sciences, Monash University, Clayton Campus, Vic. 3800, Australia.