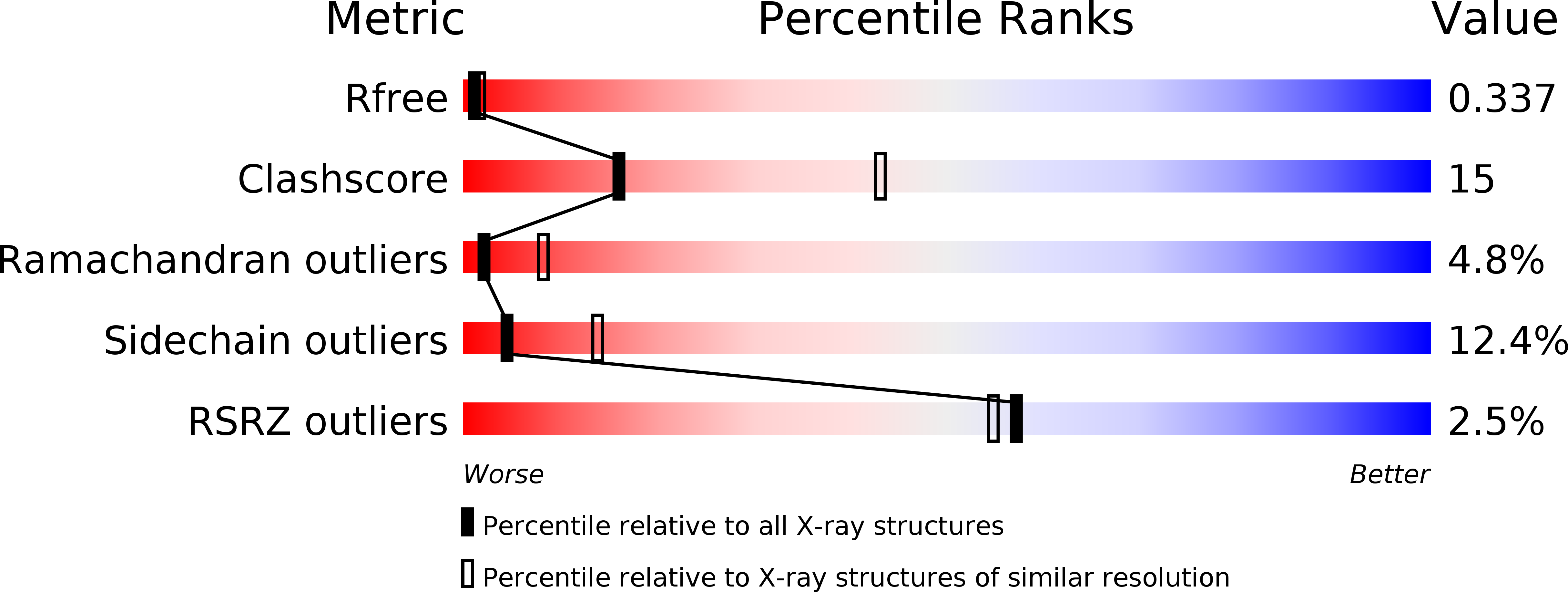
Crystal structure of an archaeal actin homolog
Roeben, A., Kofler, C., Nagy, I., Nickell, S., Ulrich Hartl, F., Bracher, A.(2006) J Mol Biol 358: 145-156
- PubMed: 16500678
- DOI: https://doi.org/10.1016/j.jmb.2006.01.096
- Primary Citation of Related Structures:
2FSJ, 2FSK, 2FSN - PubMed Abstract:
Prokaryotic homologs of the eukaryotic structural protein actin, such as MreB and ParM, have been implicated in determination of bacterial cell shape, and in the segregation of genomic and plasmid DNA. In contrast to these bacterial actin homologs, little is known about the archaeal counterparts. As a first step, we expressed a predicted actin homolog of the thermophilic archaeon Thermoplasma acidophilum, Ta0583, and determined its crystal structure at 2.1A resolution. Ta0583 is expressed as a soluble protein in T.acidophilum and is an active ATPase at physiological temperature. In vitro, Ta0583 forms sheets with spacings resembling the crystal lattice, indicating an inherent propensity to form filamentous structures. The fold of Ta0583 contains the core structure of actin and clearly belongs to the actin/Hsp70 superfamily of ATPases. Ta0583 is approximately equidistant from actin and MreB on the structural level, and combines features from both eubacterial actin homologs, MreB and ParM. The structure of Ta0583 co-crystallized with ADP indicates that the nucleotide binds at the interface between the subdomains of Ta0583 in a manner similar to that of actin. However, the conformation of the nucleotide observed in complex with Ta0583 clearly differs from that in complex with actin, but closely resembles the conformation of ParM-bound nucleotide. On the basis of sequence and structural homology, we suggest that Ta0583 derives from a ParM-like actin homolog that was once encoded by a plasmid and was transferred into a common ancestor of Thermoplasma and Ferroplasma. Intriguingly, both genera are characterized by the lack of a cell wall, and therefore Ta0583 could have a function in cellular organization.
Organizational Affiliation:
Department of Cellular Biochemistry, Max-Planck-Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany.