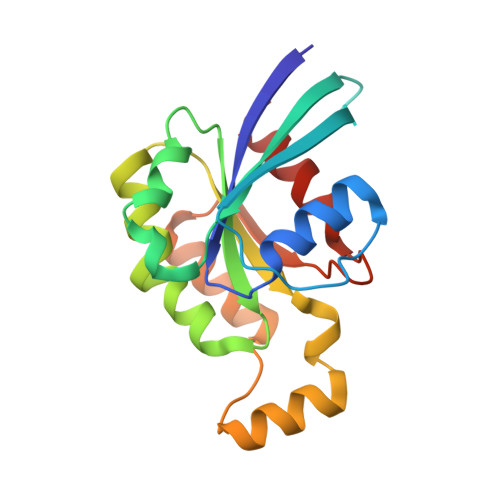
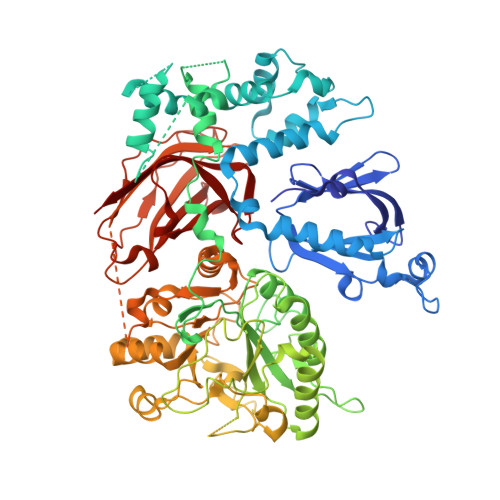
Crystal structure of Rac1 bound to its effector phospholipase C-beta2.
Jezyk, M.R., Snyder, J.T., Gershberg, S., Worthylake, D.K., Harden, T.K., Sondek, J.(2006) Nat Struct Mol Biol 13: 1135-1140
- PubMed: 17115053
- DOI: https://doi.org/10.1038/nsmb1175
- Primary Citation of Related Structures:
2FJU - PubMed Abstract:
Although diverse signaling cascades require the coordinated regulation of heterotrimeric G proteins and small GTPases, these connections remain poorly understood. We present the crystal structure of the GTPase Rac1 bound to phospholipase C-beta2 (PLC-beta2), a classic effector of heterotrimeric G proteins. Rac1 engages the pleckstrin-homology (PH) domain of PLC-beta2 to optimize its orientation for substrate membranes. Gbetagamma also engages the PH domain to activate PLC-beta2, and these two activation events are compatible, leading to additive stimulation of phospholipase activity. In contrast to PLC-delta, the PH domain of PLC-beta2 cannot bind phosphoinositides, eliminating this mode of regulation. The structure of the Rac1-PLC-beta2 complex reveals determinants that dictate selectivity of PLC-beta isozymes for Rac GTPases over other Rho-family GTPases, and substitutions within PLC-beta2 abrogate its stimulation by Rac1 but not by Gbetagamma, allowing for functional dissection of this integral signaling node.
Organizational Affiliation:
Department of Biochemistry and Biophysics The University of North Carolina School of Medicine, Chapel Hill, North Carolina 27599, USA.