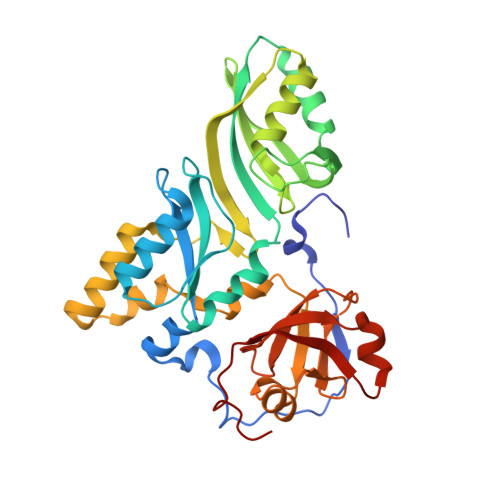
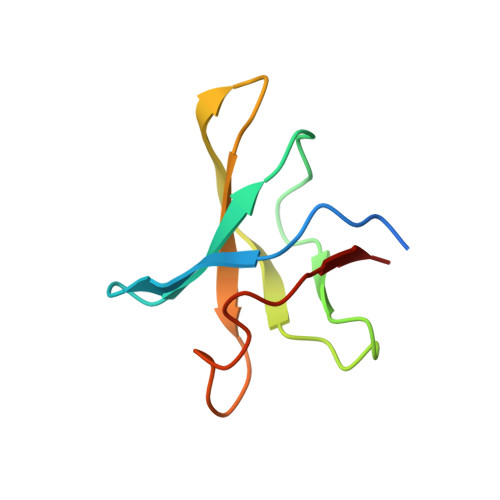
Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita.
Rashid, R., Liang, B., Baker, D.L., Youssef, O.A., He, Y., Phipps, K., Terns, R.M., Terns, M.P., Li, H.(2006) Mol Cell 21: 249-260
- PubMed: 16427014
- DOI: https://doi.org/10.1016/j.molcel.2005.11.017
- Primary Citation of Related Structures:
2EY4 - PubMed Abstract:
H/ACA RNA-protein complexes, comprised of four proteins and an H/ACA guide RNA, modify ribosomal and small nuclear RNAs. The H/ACA proteins are also essential components of telomerase in mammals. Cbf5 is the H/ACA protein that catalyzes isomerization of uridine to pseudouridine in target RNAs. Mutations in human Cbf5 (dyskerin) lead to dyskeratosis congenita. Here, we describe the 2.1 A crystal structure of a specific complex of three archaeal H/ACA proteins, Cbf5, Nop10, and Gar1. Cbf5 displays structural properties that are unique among known pseudouridine synthases and are consistent with its distinct function in RNA-guided pseudouridylation. We also describe the previously unknown structures of both Nop10 and Gar1 and the structural basis for their essential roles in pseudouridylation. By using information from related structures, we have modeled the entire ribonucleoprotein complex including both guide and substrate RNAs. We have also identified a dyskeratosis congenita mutation cluster site within a modeled dyskerin structure.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Institute of Molecular Biophysics, Florida State University, Tallahassee, Florida 32306, USA.