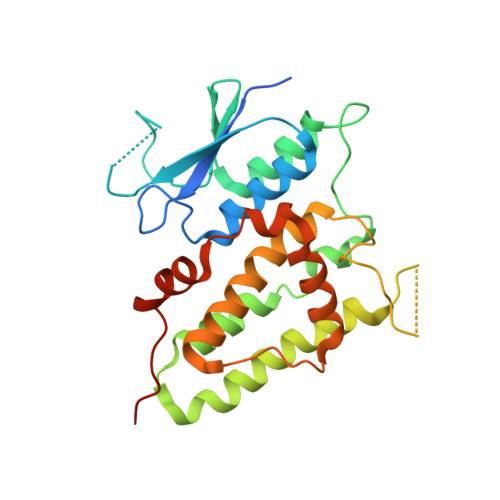
Trimeric structure of the wild soluble chloride intracellular ion channel CLIC4 observed in crystals
Li, Y.F., Li, D.F., Zeng, Z.H., Wang, D.C.(2006) Biochem Biophys Res Commun 343: 1272-1278
- PubMed: 16581025
- DOI: https://doi.org/10.1016/j.bbrc.2006.03.099
- Primary Citation of Related Structures:
2D2Z - PubMed Abstract:
The crystal structure of a wild type of the human soluble chloride intracellular ion channel CLIC4 (wCLIC4) has been determined at a resolution of 2.2A. The structure shows a homotrimer in an asymmetric unit, which is first observed in CLICs. The assembly of the trimer takes a unique triple interaction mode between three monomers with a hydrogen-bond network and hydrophobic contacts. Through such complicated interactions, the homotrimer of wCLIC4 is firmly stabilized. The structure shows an oligomeric mode with a unique assembly mechanism by which the oligomerization of CLIC4 can be performed without any intramolecular disulfide bond formation. It indicated a possibility that CLIC4 may take a unique structural organization distinct from CLIC1 for docking with lipid bilayers. In addition, the structure shows distinct conformational states of the h2 region for respective monomers of the trimer, which reveal an intrinsic conformational susceptibility for this significant region in the structural transition.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.