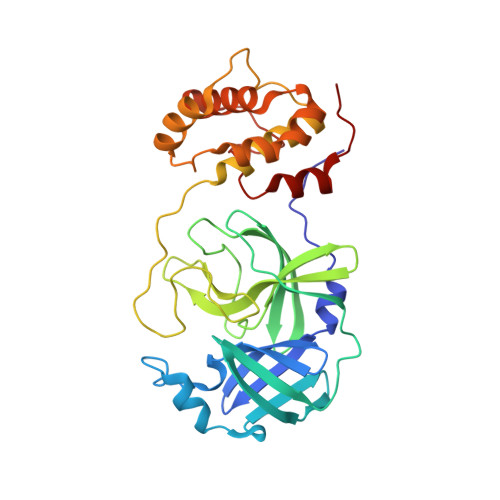
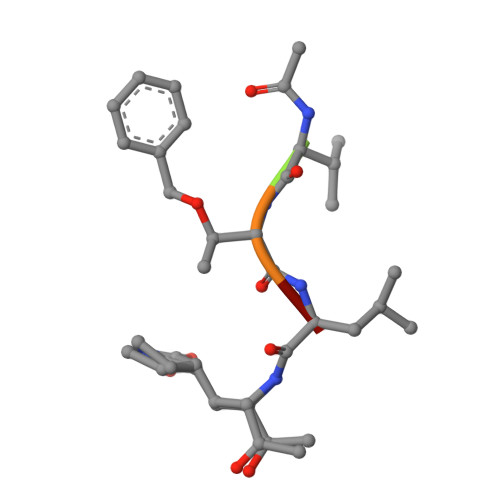
A mechanistic view of enzyme inhibition and peptide hydrolysis in the active site of the SARS-CoV 3C-like peptidase
Yin, J., Niu, C., Cherney, M.M., Zhang, J., Huitema, C., Eltis, L.D., Vederas, J.C., James, M.N.(2007) J Mol Biol 371: 1060-1074
- PubMed: 17599357
- DOI: https://doi.org/10.1016/j.jmb.2007.06.001
- Primary Citation of Related Structures:
2Z3C, 2Z3D, 2Z3E - PubMed Abstract:
The 3C-like main peptidase 3CL(pro) is a viral polyprotein processing enzyme essential for the viability of the Severe Acute Respiratory Syndrome coronavirus (SARS-CoV). While it is generalized that 3CL(pro) and the structurally related 3C(pro) viral peptidases cleave their substrates via a mechanism similar to that underlying the peptide hydrolysis by chymotrypsin-like serine proteinases (CLSPs), some of the hypothesized key intermediates have not been structurally characterized. Here, we present three crystal structures of SARS 3CL(pro) in complex with each of two members of a new class of peptide-based phthalhydrazide inhibitors. Both inhibitors form an unusual thiiranium ring with the nucleophilic sulfur atom of Cys145, trapping the enzyme's catalytic residues in configurations similar to the intermediate states proposed to exist during the hydrolysis of native substrates. Most significantly, our crystallographic data are consistent with a scenario in which a water molecule, possibly via indirect coordination from the carbonyl oxygen of Thr26, has initiated nucleophilic attack on the enzyme-bound inhibitor. Our data suggest that this structure resembles that of the proposed tetrahedral intermediate during the deacylation step of normal peptidyl cleavage.
Organizational Affiliation:
Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, AB, Canada T6G 2H7.