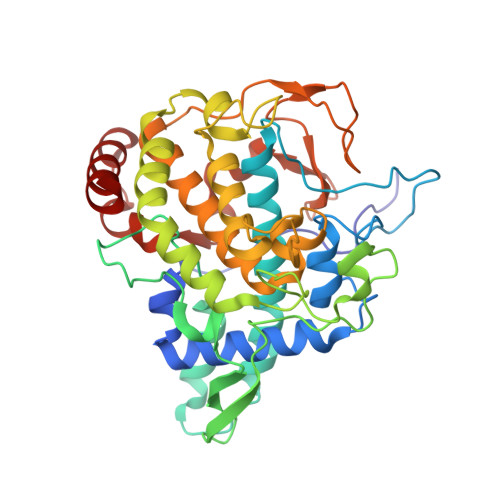
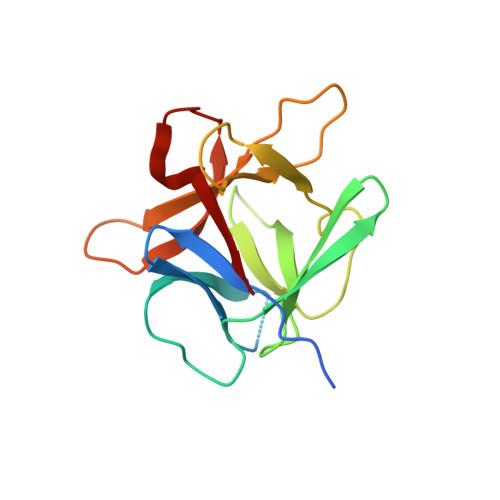
Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Ismaya, W.T., Rozeboom, H.J., Weijn, A., Mes, J.J., Fusetti, F., Wichers, H.J., Dijkstra, B.W.(2011) Biochemistry 50: 5477
- PubMed: 21598903
- DOI: https://doi.org/10.1021/bi200395t
- Primary Citation of Related Structures:
2Y9W, 2Y9X - PubMed Abstract:
Tyrosinase catalyzes the conversion of phenolic compounds into their quinone derivatives, which are precursors for the formation of melanin, a ubiquitous pigment in living organisms. Because of its importance for browning reactions in the food industry, the tyrosinase from the mushroom Agaricus bisporus has been investigated in depth. In previous studies the tyrosinase enzyme complex was shown to be a H(2)L(2) tetramer, but no clues were obtained of the identities of the subunits, their mode of association, and the 3D structure of the complex. Here we unravel this tetramer at the molecular level. Its 2.3 Å resolution crystal structure is the first structure of the full fungal tyrosinase complex. The complex comprises two H subunits of ∼392 residues and two L subunits of ∼150 residues. The H subunit originates from the ppo3 gene and has a fold similar to other tyrosinases, but it is ∼100 residues larger. The L subunit appeared to be the product of orf239342 and has a lectin-like fold. The H subunit contains a binuclear copper-binding site in the deoxy-state, in which three histidine residues coordinate each copper ion. The side chains of these histidines have their orientation fixed by hydrogen bonds or, in the case of His85, by a thioether bridge with the side chain of Cys83. The specific tyrosinase inhibitor tropolone forms a pre-Michaelis complex with the enzyme. It binds near the binuclear copper site without directly coordinating the copper ions. The function of the ORF239342 subunits is not known. Carbohydrate binding sites identified in other lectins are not conserved in ORF239342, and the subunits are over 25 Å away from the active site, making a role in activity unlikely. The structures explain how calcium ions stabilize the tetrameric state of the enzyme.
Organizational Affiliation:
Laboratory of Biophysical Chemistry, University of Groningen, Groningen, The Netherlands.