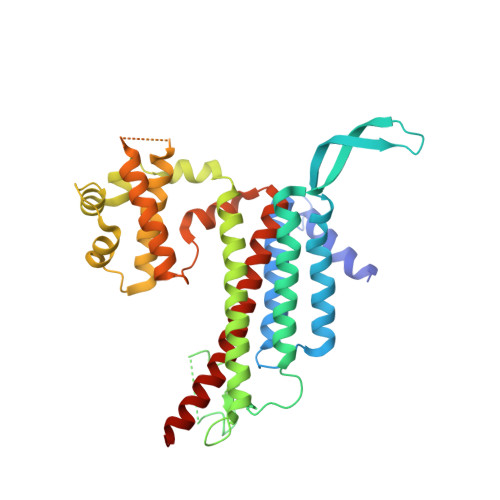
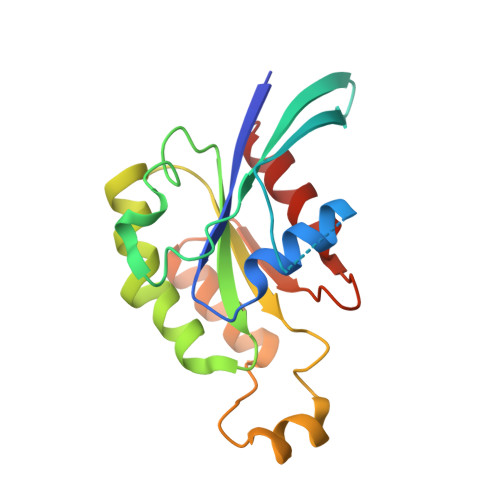
3D Structure of a Binary Rop-Prone Complex: The Final Intermediate for a Complete Set of Molecular Snapshots of the Ropgef Reaction.
Thomas, C., Fricke, I., Weyand, M., Berken, A.(2009) Biol Chem 390: 427
- PubMed: 19335195
- DOI: https://doi.org/10.1515/BC.2009.049
- Primary Citation of Related Structures:
2WBL - PubMed Abstract:
Guanine nucleotide exchange factors (GEFs) catalyze the activation of GTP-binding proteins (G proteins) in a multi-step reaction comprising intermediary complexes with and without nucleotide. Rho proteins of plants (ROPs) are activated by novel RopGEFs with a catalytic PRONE domain. We have previously characterized structures of GDP-bound ROP and a ternary complex between plant-specific ROP nucleotide exchanger (PRONE) and ROP including loosely bound GDP. Now, we complete the molecular snapshots of the RopGEF reaction with the nucleotide-free ROP-PRONE structure at 2.9 A. The binary complex surprisingly closely resembles the preceding ternary intermediate including an unusually intact P-loop in the G protein. A striking difference is the prominent contact of the invariant P-loop lysine to a conserved switch II glutamate in ROP, favoring a key role of this interaction in driving out the nucleotide. The nucleotide-free state is supported by additional interactions involving the essential WW-motif in PRONE. We propose that this GEF region stabilizes the intact P-loop conformation, which facilitates re-association with a new nucleotide and further promotes the overall exchange reaction. With our novel structure, we provide further insights into the nucleotide exchange mechanism and present a first example of the complete GEF reaction at a molecular level.
Organizational Affiliation:
Department of Structural Biology, Max Planck Institute of Molecular Physiology, Otto-Hahn-Strasse 11, D-44227 Dortmund, Germany.