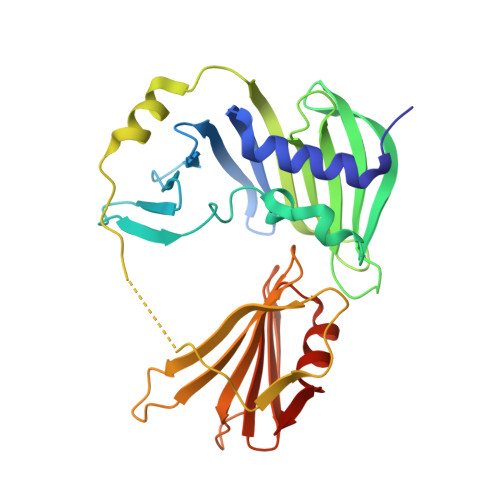
The Structure of Rseb: A Sensor in Periplasmic Stress Response of E. Coli.
Wollmann, P., Zeth, K.(2007) J Mol Biol 372: 927
- PubMed: 17692869
- DOI: https://doi.org/10.1016/j.jmb.2007.06.039
- Primary Citation of Related Structures:
2V42, 2V43 - PubMed Abstract:
An elegant network of signal transduction has evolved in the bacterial cell envelope to respond to environmental stress. It is initiated by sensing unfavourable and harmful changes in the periplasm. The stress signal is then transmitted by a controlled degradation of the transmembrane anti-sigma-factor RseA that leads to the activation of the alternative sigma factor sigma(E). The periplasmic protein RseB exerts a crucial role in modulating the stability of RseA. RseB from Escherichia coli has been crystallized and crystal structures were determined at 2.4 A and at 2.8 A resolution. The protein forms a homodimer, with the monomer composed of two domains. The large domain resembles an unclosed beta-barrel that is structurally remarkably similar to a protein family capable of binding the lipid anchor of lipoproteins. The small C-terminal domain, connected to the large domain by a partially unstructured loop, is responsible for interaction with RseA. On the basis of the structure of RseB, we suggest that it acts as a sensor of periplasmic stress with a dual functionality: it detects mislocalized lipoproteins and propagates the signal to induce the sigma(E)-response.
Organizational Affiliation:
Max Planck Institute of Biochemistry, Department of Membrane Biochemistry, Am Klopferspitz 18, D-82152 Martinsried, Germany.