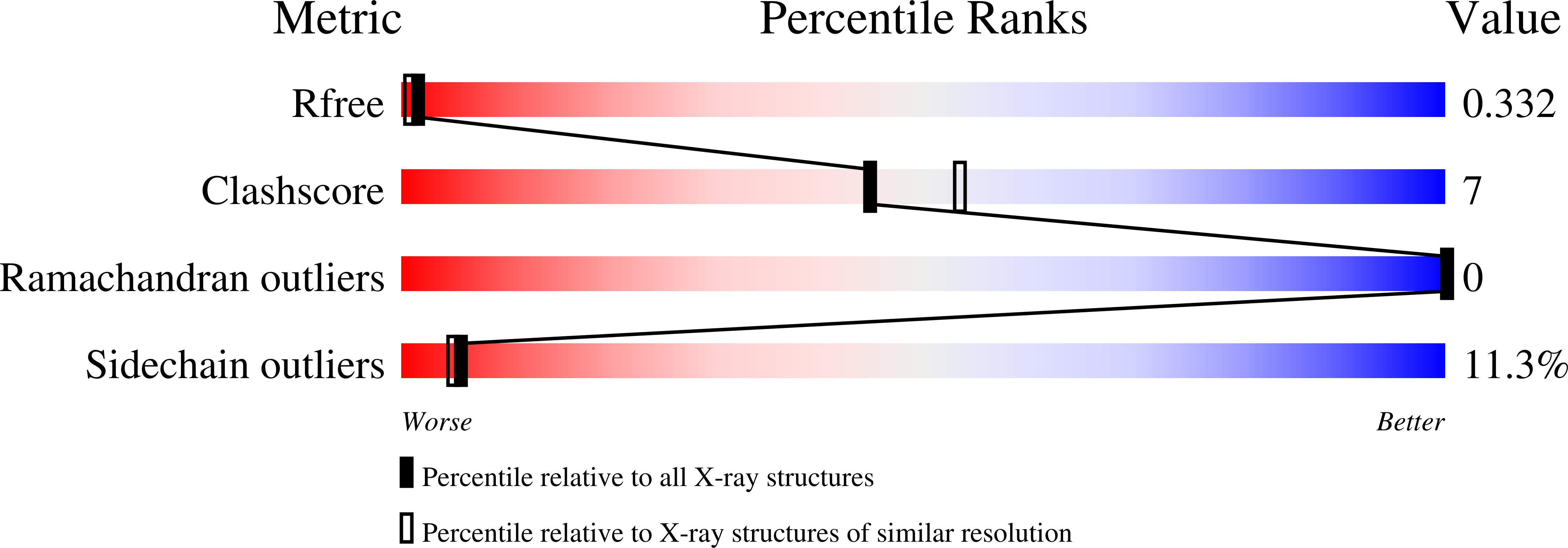
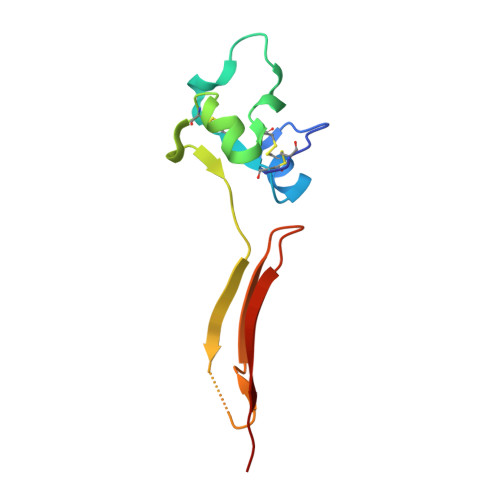
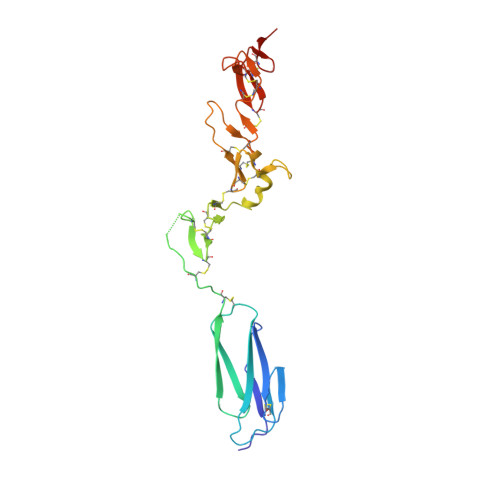
A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta2 integrins
Shi, M., Foo, S.Y., Tan, S.M., Mitchell, E.P., Law, S.K.A., Lescar, J.(2007) J Biol Chem 282: 30198-30206
- PubMed: 17673459
- DOI: https://doi.org/10.1074/jbc.M701670200
- Primary Citation of Related Structures:
2P26, 2P28 - PubMed Abstract:
Integrins mediate cell adhesion in response to activation signals that trigger conformational changes within their ectodomain. It is thought that a compact bent conformation of the molecule represents its physiological low affinity state and extended conformations its active state. We have determined the structure of two integrin fragments of the beta2 subunit. The first structure, consisting of the plexin-semaphorin-integrin domain, hybrid, integrin-epidermal growth factor 1 (I-EGF1), and I-EGF2 domains (PHE2), showed an L-shaped conformation with the bend located between the I-EGF1 and I-EGF2 domains. The second structure, which includes, in addition, the I-EGF3 domain, showed an extended conformation. The major reorientation of I-EGF2 with respect to the other domains in the two structures is accompanied by a change of torsion angle of the disulfide bond between Cys(461)-Cys(492) by 180 degrees and the conversion of a short alpha-helix (residues Ser(468)-Cys(475)) into a flexible coil. Based on the PHE2 structure, we introduced a disulfide bond between the plexin-semaphorin-integrin domain and I-EGF2 domains in the beta2 subunit. The resultant alphaLbeta2 integrin (leukocyte function-associated antigen-1) variant was locked in a bent state and could not be detected with the monoclonal antibody KIM127 in Mg(2+)/EGTA. However, it retained the binding activity to ICAM-1. These results provide a structural hypothesis for our understanding of the transition between the resting and active states of leukocyte function-associated antigen-1.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore, Singapore 637551.