Structural basis for autoinhibition of Notch
Gordon, W.R., Vardar-Ulu, D., Histen, G., Sanchez-Irizarry, C., Aster, J.C., Blacklow, S.C.(2007) Nat Struct Mol Biol 14: 295-300
- PubMed: 17401372
- DOI: https://doi.org/10.1038/nsmb1227
- Primary Citation of Related Structures:
2OO4 - PubMed Abstract:
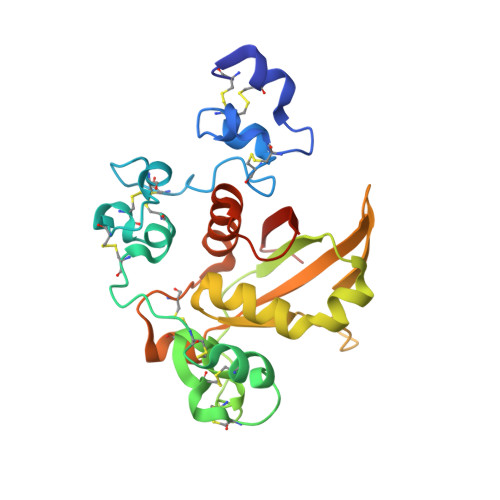
Notch receptors transmit signals between adjacent cells. Signaling is initiated when ligand binding induces metalloprotease cleavage of Notch within an extracellular negative regulatory region (NRR). We present here the X-ray structure of the human NOTCH2 NRR, which adopts an autoinhibited conformation. Extensive interdomain interactions within the NRR bury the metalloprotease site, showing that a substantial conformational movement is necessary to expose this site during activation by ligand. Leukemia-associated mutations in NOTCH1 probably release autoinhibition by destabilizing the conserved hydrophobic core of the NRR.
Organizational Affiliation:
Department of Pathology, Brigham and Women's Hospital and Harvard Medical School, 77 Ave. Louis Pasteur, Boston, Massachusetts 02115, USA.