Role of Aromatic Amino Acids in Lipopolysaccharide and Membrane Interactions of Antimicrobial Peptides for Use in Plant Disease Control.
Datta, A., Bhattacharyya, D., Singh, S., Ghosh, A., Schmidtchen, A., Malmsten, M., Bhunia, A.(2016) J Biol Chem 291: 13301-13317
- PubMed: 27137928
- DOI: https://doi.org/10.1074/jbc.M116.719575
- Primary Citation of Related Structures:
2NAT, 2NAU - PubMed Abstract:
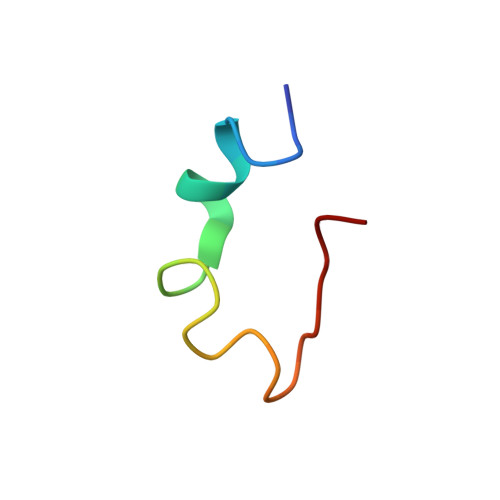
KYE28 (KYEITTIHNLFRKLTHRLFRRNFGYT-LR), the representative sequence of helix D of heparin co-factor II, was demonstrated to be potent against agronomically important Gram-negative plant pathogens Xanthomonas vesicatoria and Xanthomonas oryzae, capable of inhibiting disease symptoms in detached tomato leaves. NMR studies in the presence of lipopolysaccharide provided structural insights into the mechanisms underlying this, notably in relationship to outer membrane permeabilization. The three-dimensional solution structure of KYE28 in LPS is characterized by an N-terminal helical segment, an intermediate loop followed by another short helical stretch, and an extended C terminus. The two termini are in close proximity to each other via aromatic packing interactions, whereas the positively charged residues form an exterior polar shell. To further demonstrate the importance of the aromatic residues for this, a mutant peptide KYE28A, with Ala substitutions at Phe(11), Phe(19), Phe(23), and Tyr(25) was designed, which showed attenuated antimicrobial activity at high salt concentrations, as well as lower membrane disruption and LPS binding abilities compared with KYE28. In contrast to KYE28, KYE28A adopted an extended helical structure in LPS with extended N and C termini. Aromatic packing interactions were completely lost, although hydrophobic interaction between the side chains of hydrophobic residues were still partly retained, imparting an amphipathic character and explaining its residual antimicrobial activity and LPS binding as observed from ellipsometry and isothermal titration calorimetry. We thus present key structural aspects of KYE28, constituting an aromatic zipper, of potential importance for the development of novel plant protection agents and therapeutic agents.
Organizational Affiliation:
From the Department of Biophysics, Bose Institute, P-1/12 CIT Scheme VII (M), Kolkata 700054, India.