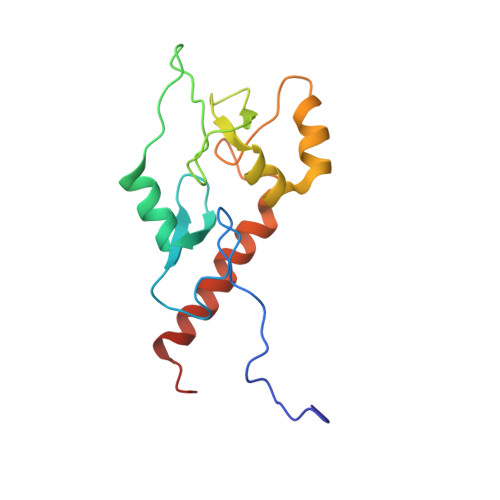
Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin
Eustermann, S., Yang, J., Law, M.J., Amos, R., Chapman, L.M., Jelinska, C., Garrick, D., Clynes, D., Gibbons, R.J., Rhodes, D., Higgs, D.R., Neuhaus, D.(2011) Nat Struct Mol Biol
- PubMed: 21666677
- DOI: https://doi.org/10.1038/nsmb.2070
- Primary Citation of Related Structures:
2LBM - PubMed Abstract:
Accurate read-out of chromatin modifications is essential for eukaryotic life. Mutations in the gene encoding X-linked ATRX protein cause a mental-retardation syndrome, whereas wild-type ATRX protein targets pericentric and telomeric heterochromatin for deposition of the histone variant H3.3 by means of a largely unknown mechanism. Here we show that the ADD domain of ATRX, in which most syndrome-causing mutations occur, engages the N-terminal tail of histone H3 through two rigidly oriented binding pockets, one for unmodified Lys4 and the other for di- or trimethylated Lys9. In vivo experiments show this combinatorial readout is required for ATRX localization, with recruitment enhanced by a third interaction through heterochromatin protein-1 (HP1) that also recognizes trimethylated Lys9. The cooperation of ATRX ADD domain and HP1 in chromatin recruitment results in a tripartite interaction that may span neighboring nucleosomes and illustrates how the 'histone-code' is interpreted by a combination of multivalent effector-chromatin interactions.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.