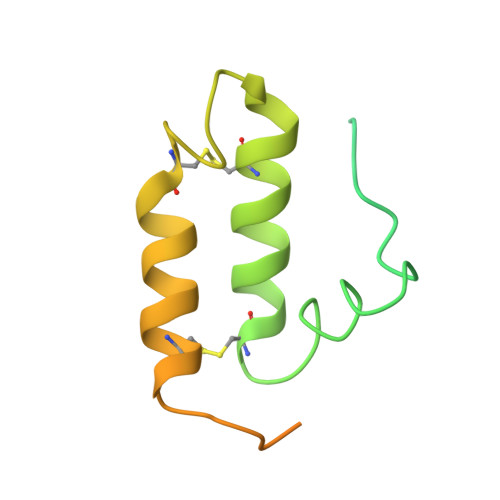
Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import.
Banci, L., Bertini, I., Cefaro, C., Cenacchi, L., Ciofi-Baffoni, S., Felli, I.C., Gallo, A., Gonnelli, L., Luchinat, E., Sideris, D., Tokatlidis, K.(2010) Proc Natl Acad Sci U S A 107: 20190-20195
- PubMed: 21059946
- DOI: https://doi.org/10.1073/pnas.1010095107
- Primary Citation of Related Structures:
2L0Y - PubMed Abstract:
Several proteins of the mitochondrial intermembrane space are targeted by internal targeting signals. A class of such proteins with α-helical hairpin structure bridged by two intramolecular disulfides is trapped by a Mia40-dependent oxidative process. Here, we describe the oxidative folding mechanism underpinning this process by an exhaustive structural characterization of the protein in all stages and as a complex with Mia40. Two consecutive induced folding steps are at the basis of the protein-trapping process. In the first one, Mia40 functions as a molecular chaperone assisting α-helical folding of the internal targeting signal of the substrate. Subsequently, in a Mia40-independent manner, folding of the second substrate helix is induced by the folded targeting signal functioning as a folding scaffold. The Mia40-induced folding pathway provides a proof of principle for the general concept that internal targeting signals may operate as a folding nucleus upon compartment-specific activation.
Organizational Affiliation:
Magnetic Resonance Center, University of Florence, Via Luigi Sacconi 6, 50019 Sesto Fiorentino, Florence, Italy. banci@cerm.unifi.it