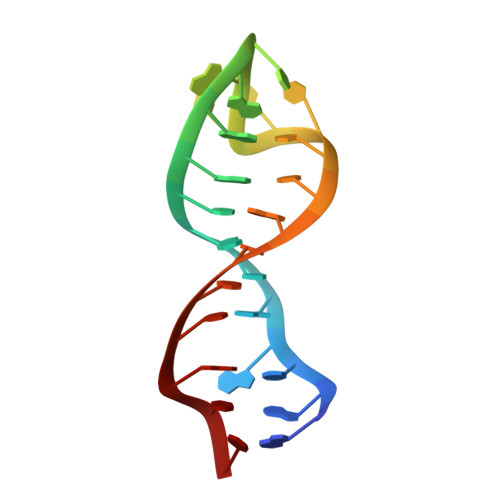
Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone).
Zheng, S., Chen, Y., Donahue, C.P., Wolfe, M.S., Varani, G.(2009) Chem Biol 16: 557-566
- PubMed: 19477420
- DOI: https://doi.org/10.1016/j.chembiol.2009.03.009
- Primary Citation of Related Structures:
2KGP - PubMed Abstract:
Some familial neurodegenerative diseases are associated with mutations that destabilize a putative stem-loop structure within an intronic region of the tau pre-messenger RNA (mRNA) and alter the production of tau protein isoforms by alternative splicing. Because stabilization of the stem loop reverses the splicing pattern associated with neurodegeneration, small molecules that stabilize this stem loop would provide new ways to dissect the mechanism of neurodegeneration and treat tauopathies. The anticancer drug mitoxantrone was recently identified in a high throughput screen to stabilize the tau pre-mRNA stem loop. Here we report the solution structure of the tau mRNA-mitoxantrone complex, validated by the structure-activity relationship of existing mitoxantrone analogs. The structure describes the molecular basis for their interaction with RNA and provides a rational basis to optimize the activity of this new class of RNA-binding molecules.
Organizational Affiliation:
Department of Chemistry, University of Washington, Seattle, 98195, USA.