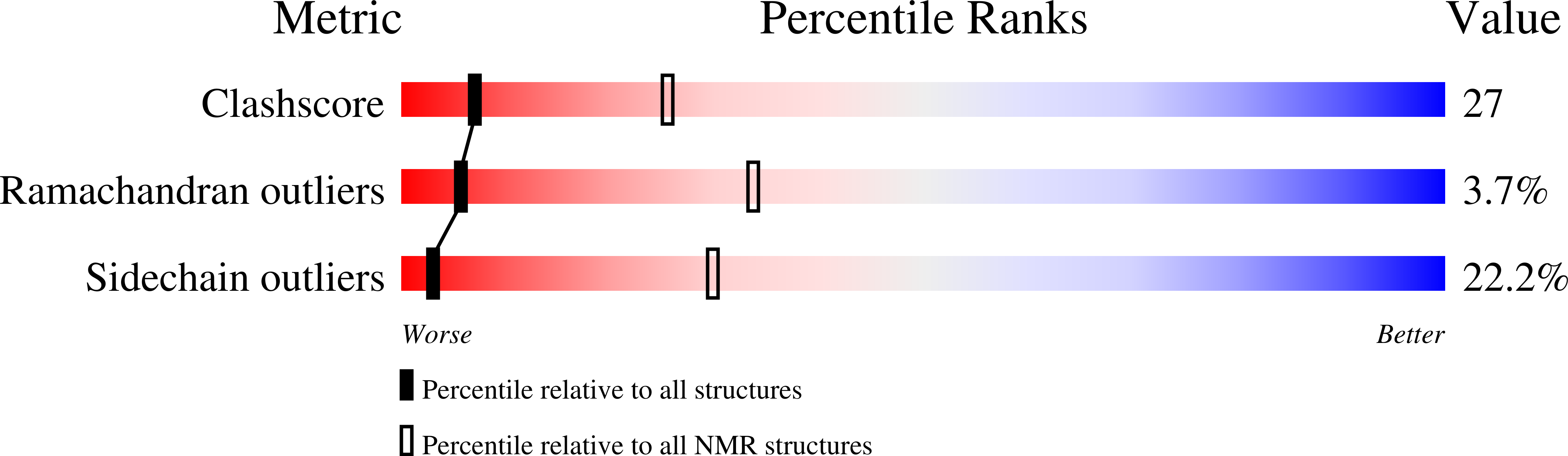Pressure-Induced Changes in the Solution Structure of the Gb1 Domain of Protein G.
Wilton, D.J., Tunnicliffe, R.B., Kamatari, Y.O., Akasaka, K., Williamson, M.P.(2008) Proteins 71: 1432
- PubMed: 18076052
- DOI: https://doi.org/10.1002/prot.21832
- Primary Citation of Related Structures:
2J52, 2J53 - PubMed Abstract:
The solution structure of the GB1 domain of protein G at a pressure of 2 kbar is presented. The structure was calculated as a change from an energy-minimised low-pressure structure using (1)H chemical shifts. Two separate changes can be characterised: a compression/distortion, which is linear with pressure; and a stabilisation of an alternative folded state. On application of pressure, linear chemical shift changes reveal that the backbone structure changes by about 0.2 A root mean square, and is compressed by about 1% overall. The alpha-helix compresses, particularly at the C-terminal end, and moves toward the beta-sheet, while the beta-sheet is twisted, with the corners closest to the alpha-helix curling up towards it. The largest changes in structure are along the second beta-strand, which becomes more twisted. This strand is where the protein binds to IgG. Curved chemical shift changes with pressure indicate that high pressure also populates an alternative structure with a distortion towards the C-terminal end of the helix, which is likely to be caused by insertion of a water molecule.
Organizational Affiliation:
Department of Molecular Biology and Biotechnology, University of Sheffield, Firth Court, Western Bank, Sheffield S10 2TN, United Kingdom.