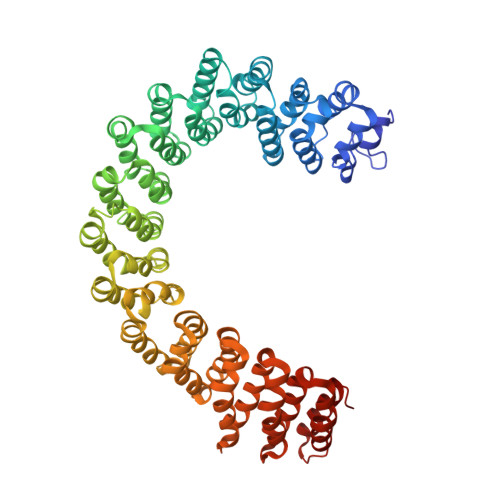Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Xing, Y., Xu, Y., Chen, Y., Jeffrey, P.D., Chao, Y., Lin, Z., Li, Z., Strack, S., Stock, J.B., Shi, Y.(2006) Cell 127: 341-353
- PubMed: 17055435
- DOI: https://doi.org/10.1016/j.cell.2006.09.025
- Primary Citation of Related Structures:
2IE3, 2IE4 - PubMed Abstract:
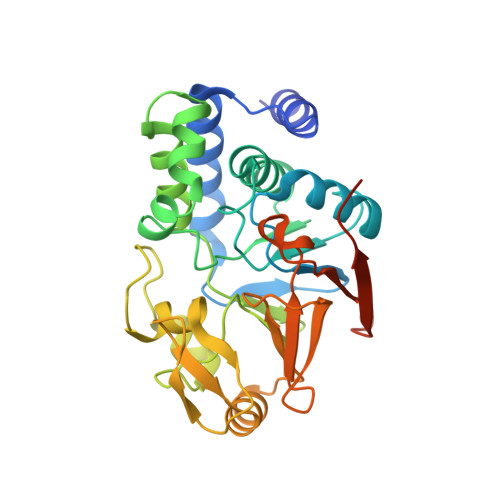
The serine/threonine phosphatase protein phosphatase 2A (PP2A) plays an essential role in many aspects of cellular functions and has been shown to be an important tumor suppressor. The core enzyme of PP2A comprises a 65 kDa scaffolding subunit and a 36 kDa catalytic subunit. Here we report the crystal structures of the PP2A core enzyme bound to two of its inhibitors, the tumor-inducing agents okadaic acid and microcystin-LR, at 2.6 and 2.8 A resolution, respectively. The catalytic subunit recognizes one end of the elongated scaffolding subunit by interacting with the conserved ridges of HEAT repeats 11-15. Formation of the core enzyme forces the scaffolding subunit to undergo pronounced structural rearrangement. The scaffolding subunit exhibits considerable conformational flexibility, which is proposed to play an essential role in PP2A function. These structures, together with biochemical analyses, reveal significant insights into PP2A function and serve as a framework for deciphering the diverse roles of PP2A in cellular physiology.
Organizational Affiliation:
Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, NJ 08544, USA.