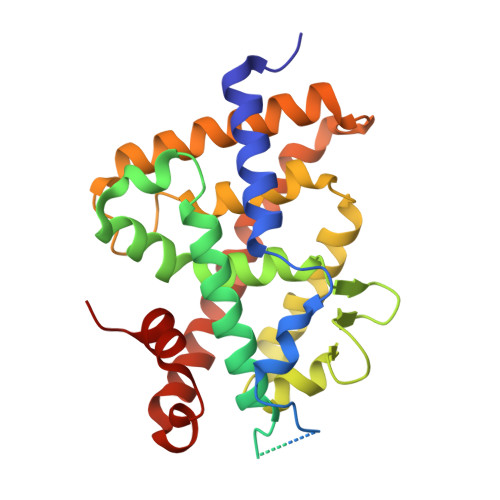
Crystal structure of the vitamin D nuclear receptor ligand binding domain in complex with a locked side chain analog of calcitriol
Rochel, N., Hourai, S., Perez-Garcia, X., Rumbo, A., Mourino, A., Moras, D.(2007) Arch Biochem Biophys 460: 172-176
- PubMed: 17346665
- DOI: https://doi.org/10.1016/j.abb.2007.01.031
- Primary Citation of Related Structures:
2HBH - PubMed Abstract:
The crystal structures of vitamin D nuclear receptor (VDR) have revealed that all compounds are anchored by the same residues to the ligand binding pocket (LBP). Based on this observation, a synthetic analog with a locked side chain (21-nor-calcitriol-20(22),23-diyne) has been synthesized in order to gain in entropy energy with a predefined active side chain conformation. The crystal structure of VDR LBD bound to this locked side chain analogue while confirming the docking provides a structural basis for the activity of this compound.
Organizational Affiliation:
Institut de Génétique et de Biologie Moléculaire et Cellulaire, Département de Biologie et de Génomique Structurales, Illkirch F-67400, France.