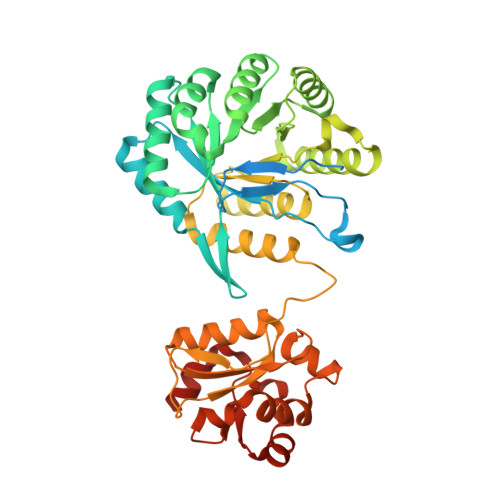
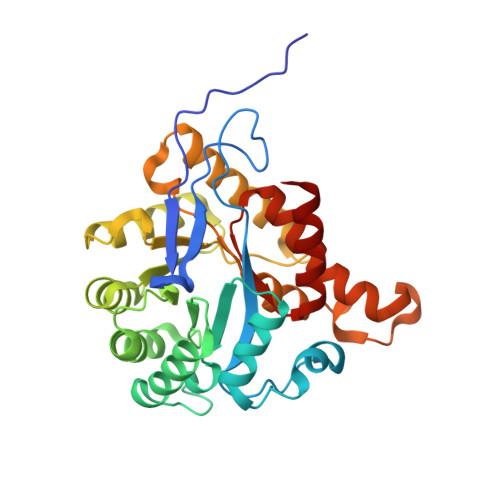
Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis.
Svetlitchnaia, T., Svetlitchnyi, V., Meyer, O., Dobbek, H.(2006) Proc Natl Acad Sci U S A 103: 14331-14336
- PubMed: 16983091
- DOI: https://doi.org/10.1073/pnas.0601420103
- Primary Citation of Related Structures:
2H9A - PubMed Abstract:
The cobalt- and iron-containing corrinoid iron-sulfur protein (CoFeSP) is functional in the acetyl-CoA (Ljungdahl-Wood) pathway of autotrophic carbon fixation in various bacteria and archaea, where it is essential for the biosynthesis of acetyl-CoA. CoFeSP acts in two methylation reactions: the transfer of a methyl group from methyltransferase (MeTr)-bound methyltetrahydrofolate to the cob(I)amide of CoFeSP and the transfer of the methyl group of methyl-cob(III)amide to the reduced Ni-Ni-[4Fe-4S] active site cluster A of acetyl-CoA synthase (ACS). We have solved the crystal structure of as-isolated CoFeSP(Ch) from the CO-oxidizing hydrogenogenic bacterium Carboxydothermus hydrogenoformans at 1.9-A resolution. The heterodimeric protein consists of two tightly interacting subunits with pseudo-twofold symmetry. The large CfsA subunit comprises three domains, of which the N-terminal domain binds the [4Fe-4S] cluster, the middle domain is a (betaalpha)(8)-barrel, and the C-terminal domain shows an open fold and binds Cobeta-aqua-(5,6-dimethylbenzimidazolylcobamide) in a "base-off" state without a protein ligand at the cobalt ion. The small CfsB subunit also displays a (betaalpha)(8)-barrel fold and interacts with the upper side of the corrin macrocycle. Structure-based alignments show that both (betaalpha)(8)-barrel domains are related to the MeTr in the acetyl-CoA pathway and to the folate domain of methionine synthase. We suggest that the C-terminal domain of the large subunit is the mobile element that allows the necessary interaction of CoFeSP(Ch) with the active site of ACS(Ch) and the methyltetrahydrofolate carrying MeTr. The conformation in the crystal structure shields the two open coordinations of cobalt and likely represents a resting state.
Organizational Affiliation:
Laboratorium Proteinkristallographie, Lehrstuhl für Mikrobiologie, and Bayreuther Zentrum für Molekulare Biowissenschaften, Universität Bayreuth, 95440 Bayreuth, Germany.