Structure, dynamics and heparin binding of the C-terminal domain of insulin-like growth factor-binding protein-2 (IGFBP-2)
Kuang, Z., Yao, S., Keizer, D.W., Wang, C.C., Bach, L.A., Forbes, B.E., Wallace, J.C., Norton, R.S.(2006) J Mol Biol 364: 690-704
- PubMed: 17020769
- DOI: https://doi.org/10.1016/j.jmb.2006.09.006
- Primary Citation of Related Structures:
2H7T - PubMed Abstract:
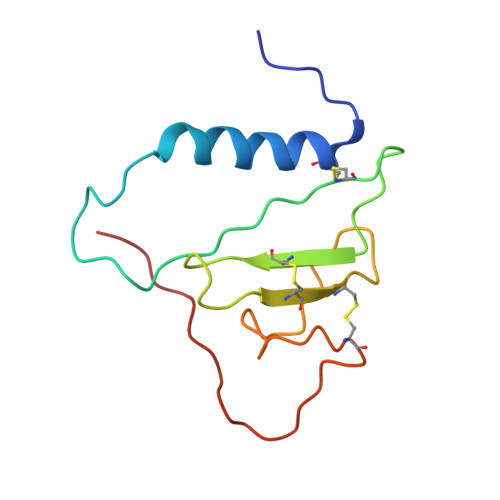
Insulin-like growth factor-binding protein-2 (IGFBP-2) is the largest member of a family of six proteins (IGFBP-1 to 6) that bind insulin-like growth factors I and II (IGF-I/II) with high affinity. In addition to regulating IGF actions, IGFBPs have IGF-independent functions. The C-terminal domains of IGFBPs contribute to high-affinity IGF binding, and confer binding specificity and have overlapping but variable interactions with many other molecules. Using nuclear magnetic resonance (NMR) spectroscopy, we have determined the solution structure of the C-terminal domain of IGFBP-2 (C-BP-2) and analysed its backbone dynamics based on 15N relaxation parameters. C-BP-2 has a thyroglobulin type 1 fold consisting of an alpha-helix, a three-stranded anti-parallel beta-sheet and three flexible loops. Compared to C-BP-6 and C-BP-1, structural differences that may affect IGF binding and underlie other functional differences were found. C-BP-2 has a longer disordered loop I, and an extended C-terminal tail, which is unstructured and very mobile. The length of the helix is identical with that of C-BP-6 but shorter than that of C-BP-1. Reduced spectral density mapping analysis showed that C-BP-2 possesses significant rapid motion in the loops and termini, and may undergo slower conformational or chemical exchange in the structured core and loop II. An RGD motif is located in a solvent-exposed turn. A pH-dependent heparin-binding site on C-BP-2 has been identified. Protonation of two histidine residues, His271 and His228, seems to be important for this binding, which occurs at slightly acidic pH (6.0) and is more significant at pH 5.5, but is largely suppressed at pH 7.4. Possible preferential binding of IGFBP-2 and its C- domain fragments to glycosaminoglycans in the acidic extracellular matrix (ECM) of tumours may be related to their roles in cancer.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville 3050, Australia.