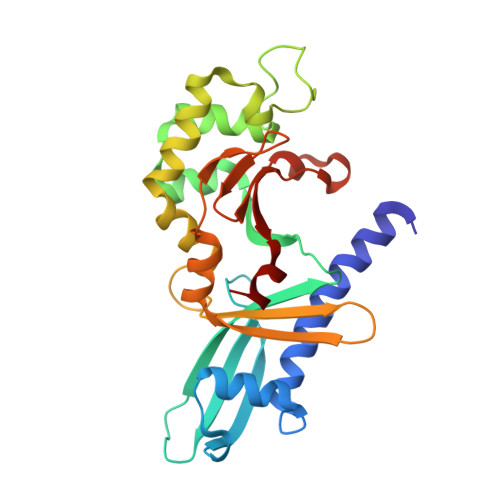
DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion.
Horton, J.R., Zhang, X., Maunus, R., Yang, Z., Wilson, G.G., Roberts, R.J., Cheng, X.(2006) Nucleic Acids Res 34: 939-948
- PubMed: 16473850
- DOI: https://doi.org/10.1093/nar/gkj484
- Primary Citation of Related Structures:
2FKC, 2FKH, 2FL3, 2FLC - PubMed Abstract:
HinP1I recognizes and cleaves the palindromic tetranucleotide sequence G downward arrowCGC in DNA. We report three structures of HinP1I-DNA complexes: in the presence of Ca(2+) (pre-reactive complex), in the absence of metal ion (binary complex) and in the presence of Mg(2+) (post-reactive complex). HinP1I forms a back-to-back dimer with two active sites and two DNA duplexes bound on the outer surfaces of the dimer facing away from each other. The 10 bp DNA duplexes undergo protein-induced distortions exhibiting features of A-, B- and Z-conformations: bending on one side (by intercalation of a phenylalanine side chain into the major groove), base flipping on the other side of the recognition site (by expanding the step rise distance of the local base pair to Z-form) and a local A-form conformation between the two central C:G base pairs of the recognition site (by binding of the N-terminal helix in the minor groove). In the pre- and post-reactive complexes, two metals (Ca(2+) or Mg(2+)) are found in the active site. The enzyme appears to cleave DNA sequentially, hydrolyzing first one DNA strand, as seen in the post-reactive complex in the crystalline state, and then the other, as supported by the observation that, in solution, a nicked DNA intermediate accumulates before linearization.
Organizational Affiliation:
Department of Biochemistry, Emory University School of Medicine 1510 Clifton Road, Atlanta, GA 30322, USA.