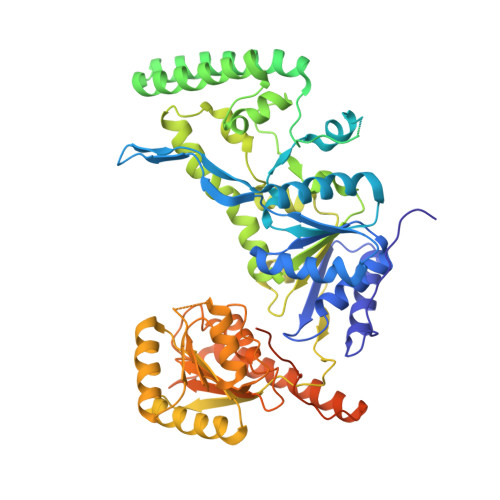
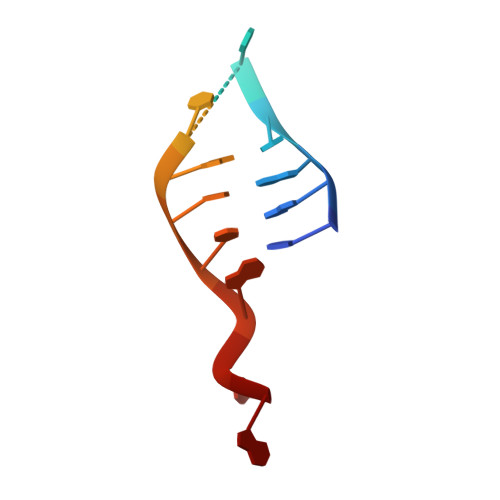
Structural basis for DNA recognition and processing by UvrB.
Truglio, J.J., Karakas, E., Rhau, B., Wang, H., DellaVecchia, M.J., Van Houten, B., Kisker, C.(2006) Nat Struct Mol Biol 13: 360-364
- PubMed: 16532007
- DOI: https://doi.org/10.1038/nsmb1072
- Primary Citation of Related Structures:
2FDC - PubMed Abstract:
DNA-damage recognition in the nucleotide excision repair (NER) cascade is a complex process, operating on a wide variety of damages. UvrB is the central component in prokaryotic NER, directly involved in DNA-damage recognition and guiding the DNA through repair synthesis. We report the first structure of a UvrB-double-stranded DNA complex, providing insights into the mechanism by which UvrB binds DNA, leading to formation of the preincision complex. One DNA strand, containing a 3' overhang, threads behind a beta-hairpin motif of UvrB, indicating that this motif inserts between the strands of the double helix, thereby locking down either the damaged or undamaged strand. The nucleotide directly behind the beta-hairpin is flipped out and inserted into a small, highly conserved pocket in UvrB.
Organizational Affiliation:
Department of Pharmacological Sciences, Stony Brook University, Stony Brook, New York 11794-5115, USA.