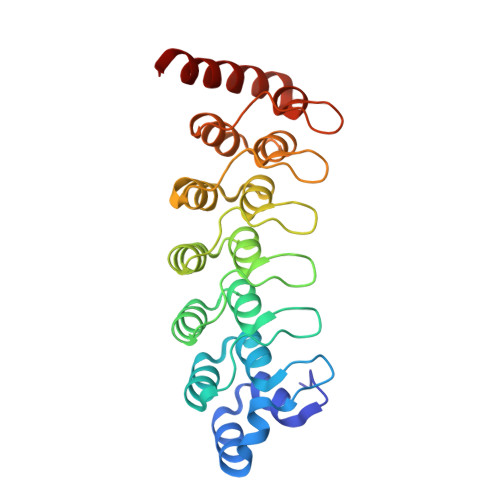
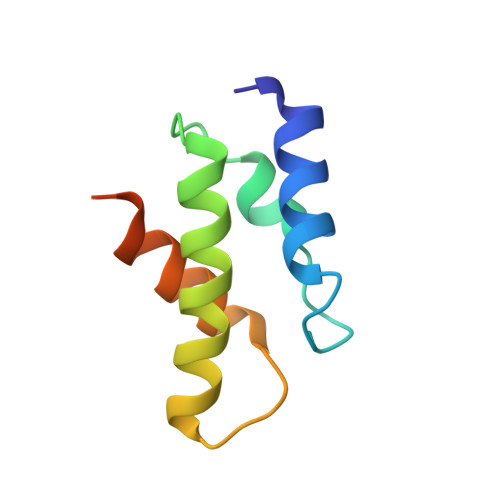
Structure of the Oncoprotein Gankyrin in Complex with S6 ATPase of the 26S Proteasome
Nakamura, Y., Nakano, K., Umehara, T., Kimura, M., Hayashizaki, Y., Tanaka, A., Horikoshi, M., Padmanabhan, B., Yokoyama, S.(2007) Structure 15: 179-189
- PubMed: 17292836
- DOI: https://doi.org/10.1016/j.str.2006.11.015
- Primary Citation of Related Structures:
2DVW - PubMed Abstract:
Gankyrin is an oncoprotein commonly overexpressed in most hepatocellular carcinomas. Gankyrin interacts with S6 ATPase of the 19S regulatory particle of the 26S proteasome and enhances the degradation of the tumor suppressors pRb and p53. Here, we report the structure of gankyrin in complex with the C-terminal domain of S6 ATPase. Almost all of the seven ankyrin repeats of gankyrin interact, through its concave region, with the C-terminal domain of S6 ATPase. The intermolecular interactions occur through the complementary charged residues between gankyrin and S6 ATPase. Biochemical studies based on the structure of the complex revealed that gankyrin interacts with pRb in both the presence and absence of S6 ATPase; however, the E182 residue in gankyrin is essential for the pRb interaction. These results provide a structural basis for the involvement of gankyrin in the pRb degradation pathway, through its association with S6 ATPase of the 26S proteasome.
Organizational Affiliation:
RIKEN Genomic Sciences Center, 1-7-22 Suehiro-cho, Tsurumi, Yokohama 230-0045, Japan.