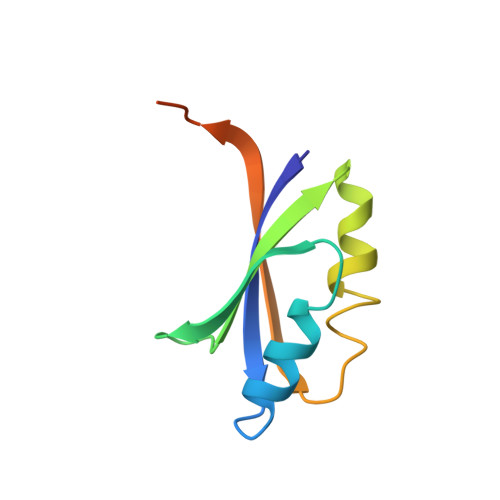
Structure of the stand-alone RAM-domain protein from Thermus thermophilus HB8
Nakano, N., Okazaki, N., Satoh, S., Takio, K., Kuramitsu, S., Shinkai, A., Yokoyama, S.(2006) Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 855-860
- PubMed: 16946463
- DOI: https://doi.org/10.1107/S1744309106031150
- Primary Citation of Related Structures:
2DJW - PubMed Abstract:
The stand-alone RAM (regulation of amino-acid metabolism) domain protein SraA from Thermus thermophilus HB8 (TTHA0845) was crystallized in the presence of zinc ions. The X-ray crystal structure was determined using a multiple-wavelength anomalous dispersion technique and was refined at 2.4 A resolution to a final R factor of 25.0%. The monomeric structure is a betaalphabetabetaalphabeta fold and it dimerizes mainly through interactions between the antiparallel beta-sheets. Furthermore, five SraA dimers form a ring with external and internal diameters of 70 and 20 A, respectively. This decameric structure is unique compared with the octameric and dodecameric structures found for other stand-alone RAM-domain proteins and the C-terminal RAM domains of Lrp/AsnC-family proteins.
Organizational Affiliation:
RIKEN SPring-8 Center, Harima Institute, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan.