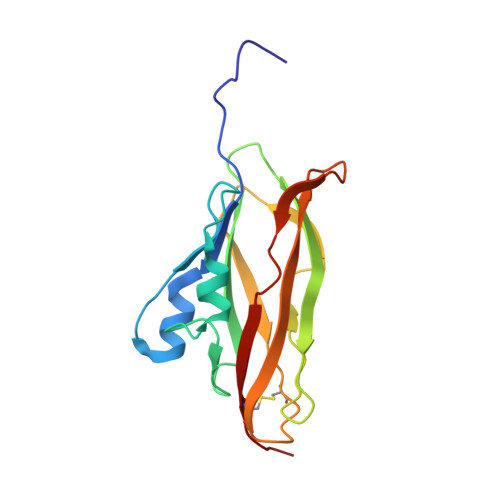
NMR structure of the WIF domain of the human Wnt-inhibitory factor-1
Liepinsh, E., Banyai, L., Patthy, L., Otting, G.(2006) J Mol Biol 357: 942-950
- PubMed: 16476441
- DOI: https://doi.org/10.1016/j.jmb.2006.01.047
- Primary Citation of Related Structures:
2D3J - PubMed Abstract:
The human Wnt-binding protein Wnt-inhibitory factor-1 (WIF-1) comprises an N-terminal WIF module followed by five EGF-like repeats. Here we report the three-dimensional structure of the WIF domain of WIF-1 determined by NMR spectroscopy. The fold consists of an eight-stranded beta-sandwich reminiscent of the immunoglobulin fold. Residual detergent (Brij-35) used in the refolding protocol was found to bind tightly to the WIF domain. The binding site was identified by intermolecular nuclear Overhauser effects observed between the WIF domain and the alkyl chain of the detergent. The results point to a possible role of WIF domains as a recognition motif of Wnt and Drosophila Hedgehog proteins that are activated by palmitoylation.
Organizational Affiliation:
Institute of Organic Synthesis, LV-1006 Riga, Latvia.