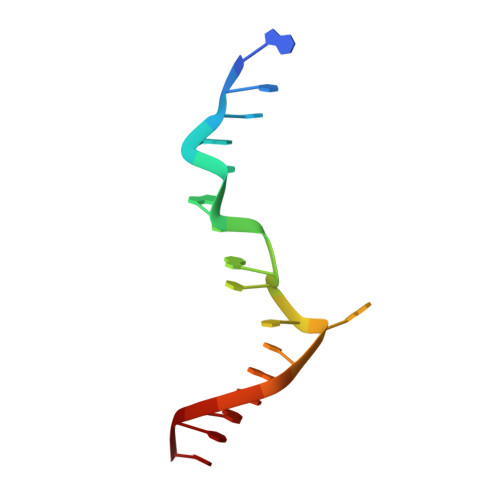
Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases.
Ha, S.C., Lowenhaupt, K., Rich, A., Kim, Y.G., Kim, K.K.(2005) Nature 437: 1183-1186
- PubMed: 16237447
- DOI: https://doi.org/10.1038/nature04088
- Primary Citation of Related Structures:
2ACJ - PubMed Abstract:
Left-handed Z-DNA is a higher-energy form of the double helix, stabilized by negative supercoiling generated by transcription or unwrapping nucleosomes. Regions near the transcription start site frequently contain sequence motifs favourable for forming Z-DNA, and formation of Z-DNA near the promoter region stimulates transcription. Z-DNA is also stabilized by specific protein binding; several proteins have been identified with low nanomolar binding constants. Z-DNA occurs in a dynamic state, forming as a result of physiological processes then relaxing to the right-handed B-DNA. Each time a DNA segment turns into Z-DNA, two B-Z junctions form. These have been examined extensively, but their structure was unknown. Here we describe the structure of a B-Z junction as revealed by X-ray crystallography at 2.6 A resolution. A 15-base-pair segment of DNA is stabilized at one end in the Z conformation by Z-DNA binding proteins, while the other end remains B-DNA. Continuous stacking of bases between B-DNA and Z-DNA segments is found, with the breaking of one base pair at the junction and extrusion of the bases on each side (Fig. 1). These extruded bases may be sites for DNA modification.
Organizational Affiliation:
Department of Molecular Cell Biology, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Suwon 440-746, Korea.