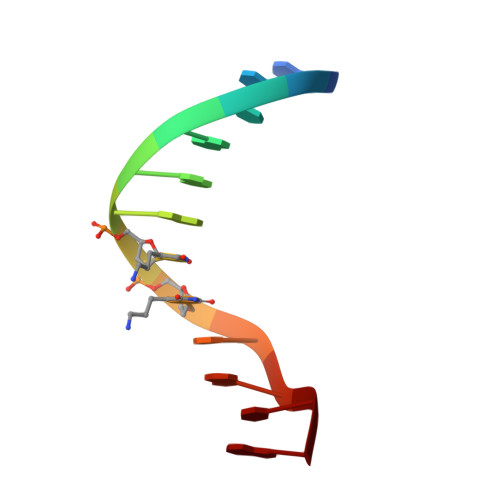
Structure of B-DNA with cations tethered in the major groove.
Moulaei, T., Maehigashi, T., Lountos, G.T., Komeda, S., Watkins, D., Stone, M.P., Marky, L.A., Li, J.S., Gold, B., Williams, L.D.(2005) Biochemistry 44: 7458-7468
- PubMed: 15895989
- DOI: https://doi.org/10.1021/bi050128z
- Primary Citation of Related Structures:
1Z5T - PubMed Abstract:
Here, we describe the 1.6-A X-ray structure of the DDD (Dickerson-Drew dodecamer), which has been covalently modified by the tethering of four cationic charges. This modified version of the DDD, called here the DDD(4+), is composed of [d(CGCGAAXXCGCG)](2), where X is effectively a thymine residue linked at the 5 position to an n-propyl-amine. The structure was determined from crystals soaked with thallium(I), which has been broadly used as a mimic of K(+) in X-ray diffraction experiments aimed at determining positions of cations adjacent to nucleic acids. Three of the tethered cations are directed radially out from the DNA. The radially directed tethered cations do not appear to induce structural changes or to displace counterions. One of the tethered cations is directed in the 3' direction, toward a phosphate group near one end of the duplex. This tethered cation appears to interact electrostatically with the DNA. This interaction is accompanied by changes in helical parameters rise, roll, and twist and by a displacement of the backbone relative to a control oligonucleotide. In addition, these interactions appear to be associated with displacement of counterions from the major groove of the DNA.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, Georgia 30332-0400, USA.