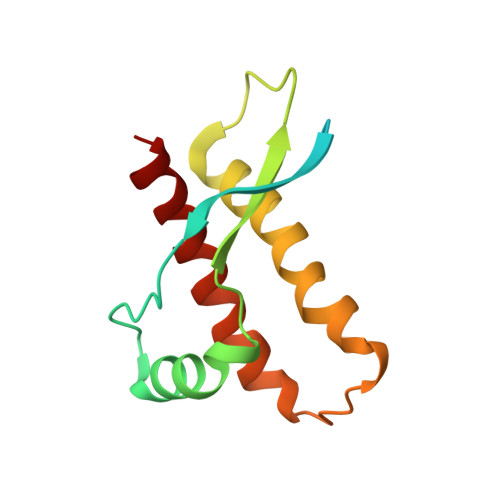
Prion protein NMR structures of chicken, turtle, and frog
Calzolai, L., Lysek, D.A., Perez, D.R., Guntert, P., Wuthrich, K.(2005) Proc Natl Acad Sci U S A 102: 651-655
- PubMed: 15647366
- DOI: https://doi.org/10.1073/pnas.0408939102
- Primary Citation of Related Structures:
1U3M, 1U5L, 1XU0 - PubMed Abstract:
The NMR structures of the recombinant prion proteins from chicken (Gallus gallus; chPrP), the red-eared slider turtle (Trachemys scripta; tPrP), and the African clawed frog (Xenopus laevis; xlPrP) are presented. The amino acid sequences of these prion proteins show approximately 30% identity with mammalian prion proteins. All three species form the same molecular architecture as mammalian PrPC, with a long, flexibly disordered tail attached to the N-terminal end of a globular domain. The globular domain in chPrP and tPrP contains three alpha-helices, one short 3(10)-helix, and a short antiparallel beta-sheet. In xlPrP, the globular domain includes three alpha-helices and a somewhat longer beta-sheet than in the other species. The spatial arrangement of these regular secondary structures coincides closely with that of the globular domain in mammalian prion proteins. Based on the low sequence identity to mammalian PrPs, comparison of chPrP, tPrP, and xlPrP with mammalian PrPC structures is used to identify a set of essential amino acid positions for the preservation of the same PrPC fold in birds, reptiles, amphibians, and mammals. There are additional conserved residues without apparent structural roles, which are of interest for the ongoing search for physiological functions of PrPC in healthy organisms.
Organizational Affiliation:
Institut für Molekularbiologie und Biophysik, Eidgenössische Technische Hochschule Zürich, CH-8093 Zürich, Switzerland.