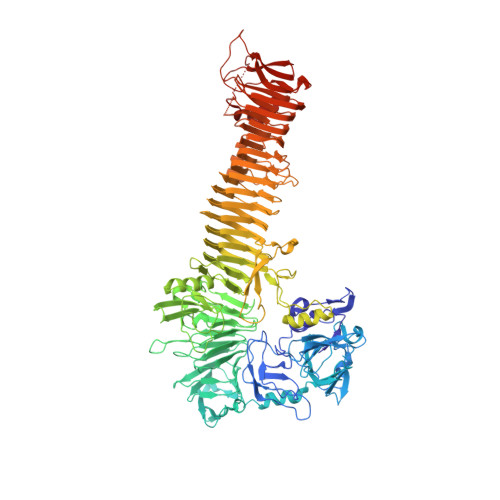
Crystal structure of heme binding protein, an autotransporter hemoglobin protease from pathogenic escherichia coli
Otto, B.R., Sijbrandi, R., Luirink, J., Oudega, B., Heddle, J.G., Mizutani, K., Park, S.-Y., Tame, J.R.H.(2005) J Biol Chem 280: 17339-17345
- PubMed: 15728184
- DOI: https://doi.org/10.1074/jbc.M412885200
- Primary Citation of Related Structures:
1WXR - PubMed Abstract:
The acquisition of iron is essential for the survival of pathogenic bacteria, which have consequently evolved a wide variety of uptake systems to extract iron and heme from host proteins such as hemoglobin. Hemoglobin protease (Hbp) was discovered as a factor involved in the symbiosis of pathogenic Escherichia coli and Bacteroides fragilis, which cause intra-abdominal abscesses. Released from E. coli, this serine protease autotransporter degrades hemoglobin and delivers heme to both bacterial species. The crystal structure of the complete passenger domain of Hbp (110 kDa) is presented, which is the first structure from this class of serine proteases and the largest parallel beta-helical structure yet solved.
Organizational Affiliation:
Department of Molecular Microbiology, Faculty of Earth and Life Sciences, Vrije Universiteit de Boelelaan 1085, 1081 HV Amsterdam, The Netherlands.