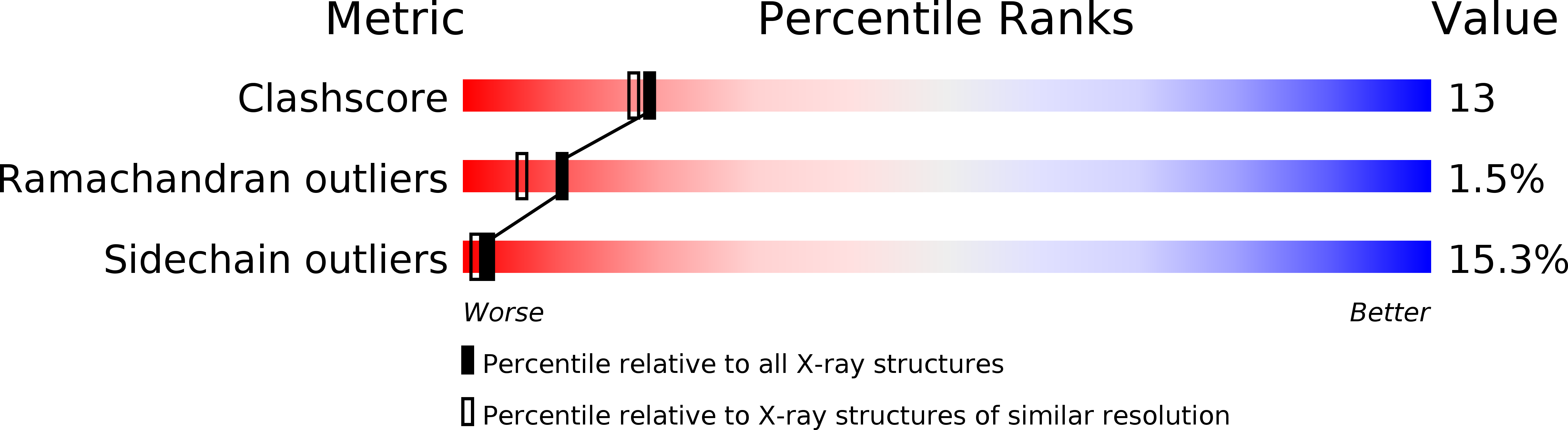Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Oikonomakos, N.G., Kosmopoulou, M.N., Chrysina, E.D., Leonidas, D.D., Kostas, I.D., Wendt, K.U., Klabunde, T., Defossa, E.(2005) Protein Sci 14: 1760-1771
- PubMed: 15987904
- DOI: https://doi.org/10.1110/ps.051432405
- Primary Citation of Related Structures:
1WUT, 1WUY, 1WV0, 1WV1 - PubMed Abstract:
Acyl ureas were discovered as a novel class of inhibitors for glycogen phosphorylase, a molecular target to control hyperglycemia in type 2 diabetics. This series is exemplified by 6-{2,6-Dichloro- 4-[3-(2-chloro-benzoyl)-ureido]-phenoxy}-hexanoic acid, which inhibits human liver glycogen phosphorylase a with an IC(50) of 2.0 microM. Here we analyze four crystal structures of acyl urea derivatives in complex with rabbit muscle glycogen phosphorylase b to elucidate the mechanism of inhibition of these inhibitors. The structures were determined and refined to 2.26 Angstroms resolution and demonstrate that the inhibitors bind at the allosteric activator site, where the physiological activator AMP binds. Acyl ureas induce conformational changes in the vicinity of the allosteric site. Our findings suggest that acyl ureas inhibit glycogen phosphorylase by direct inhibition of AMP binding and by indirect inhibition of substrate binding through stabilization of the T' state.
Organizational Affiliation:
Institute of Organic and Pharmaceutical Chemistry, The National Hellenic Research Foundation, Athens, Greece. ngo@eie.gr