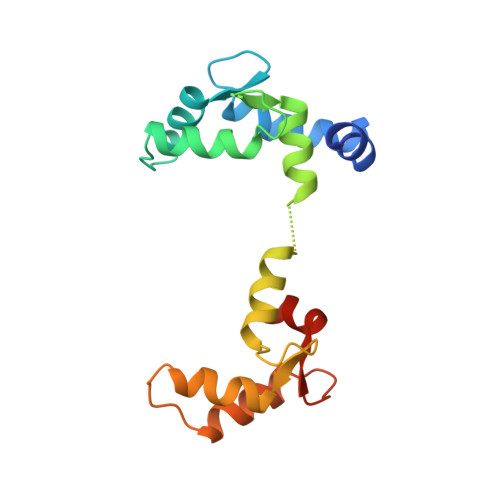
NMR solution structure of calcium-saturated skeletal muscle troponin C.
Slupsky, C.M., Sykes, B.D.(1995) Biochemistry 34: 15953-15964
- PubMed: 8519752
- DOI: https://doi.org/10.1021/bi00049a010
- Primary Citation of Related Structures:
1TNW, 1TNX - PubMed Abstract:
Troponin C (TnC) is an 18 kDa (162-residue) thin-filament calcium-binding protein responsible for triggering muscle contraction upon the release of calcium from the sarcoplasmic reticulum. The structure of TnC with two calcium ions bound has previously been solved by X-ray methods. Shown here is the solution structure of TnC which has been solved using 3D and 4D heteronuclear nuclear magnetic resonance (NMR) spectroscopic techniques. The 1H, 13C, and 15N backbone chemical shifts have already been published [Slupsky, C. M., Reinach, F. C., Smillie, L. B., & Sykes, B. D. (1995) Protein Sci. 4, 1279-1290]. Presented herein are the 1H, 13C, and 15N side-chain chemical shifts which are 80% complete. The structure of calcium-saturated TnC was determined on the basis of 2106 NOE-derived distance restraints, 121 phi dihedral angle restraints, and 76 psi dihedral angle restraints. The appearance of calcium-saturated TnC reveals a dumbbell-shaped molecule with two globular domains connected by a linker. The structures of the N-terminal and C-terminal domains are highly converged [backbone atomic root mean square deviations (rmsd) about the mean atomic coordinate position for residues 10-80 and 98-155 are 0.66 +/- 0.17 and 0.69 +/- 0.18 A, respectively]; however, the orientation of one domain with respect to the other is not well-defined, and thus each domain appears to be structurally independent. Comparison of the calcium-saturated form of TnC determined herein with the half-saturated form determined by X-ray methods reveals two major differences. First, there is a major structural change which occurs in the N-terminal domain resulting in the opening of a hydrophobic pocket presumably to present itself to its target protein troponin I. This structural change appears to involve only helices B and C which move away from helices N/A/D by the alteration of the backbone phi, psi angles of glutamic acid 41 from irregular in the crystal structure (-97 degrees, -7 degrees) to helical in the NMR calcium-saturated structure (-60 degrees, -34 degrees). The other difference between the two structures is the presence of a flexible linker between the two domains in the NMR structure. This flexible linker allows the two domains of TnC to adopt any orientation with respect to one another such that they can interact with a variety of targets.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, Canada.