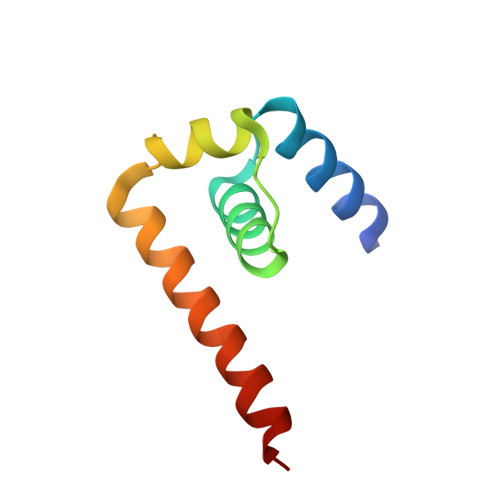
West nile virus core protein; tetramer structure and ribbon formation
Dokland, T., Walsh, M., Mackenzie, J.M., Khromykh, A.A., Ee, K.-H., Wang, S.(2004) Structure 12: 1157-1163
- PubMed: 15242592
- DOI: https://doi.org/10.1016/j.str.2004.04.024
- Primary Citation of Related Structures:
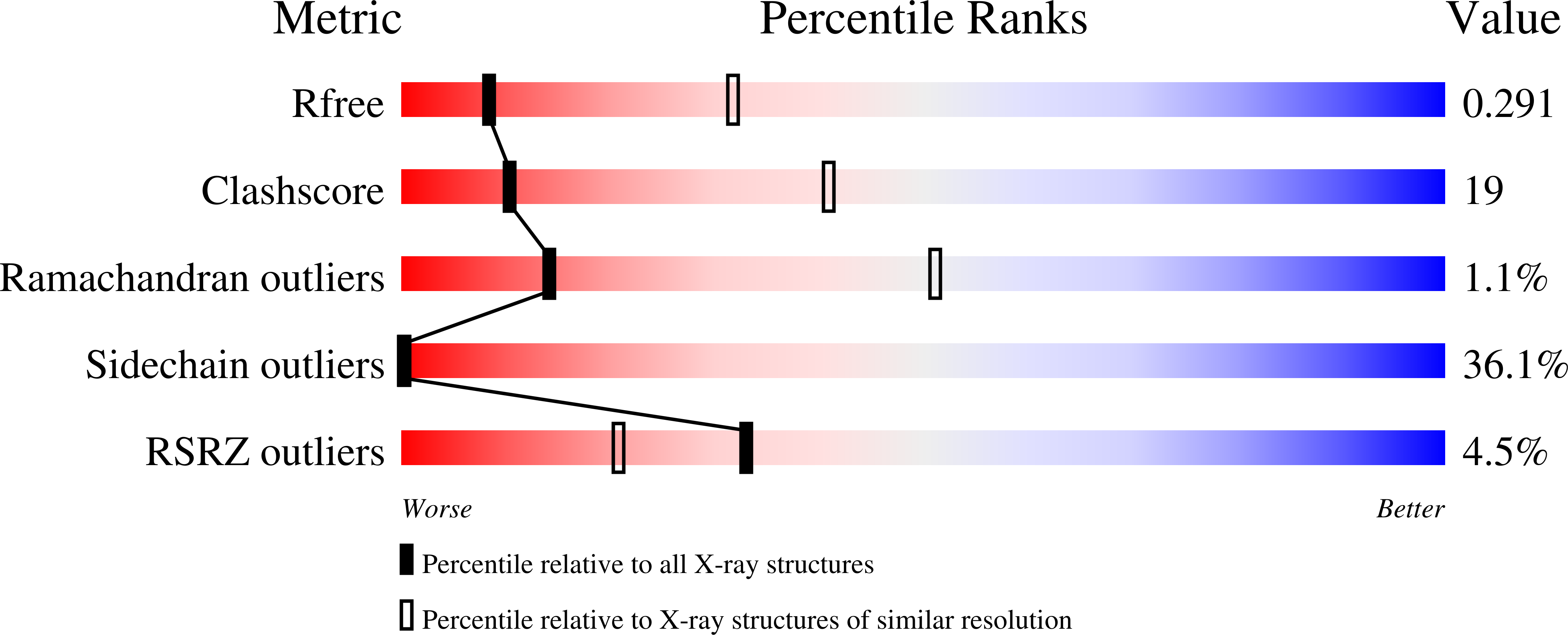
1SFK - PubMed Abstract:
We have determined the crystal structure of the core (C) protein from the Kunjin subtype of West Nile virus (WNV), closely related to the NY99 strain of WNV, currently a major health threat in the U.S. WNV is a member of the Flaviviridae family of enveloped RNA viruses that contains many important human pathogens. The C protein is associated with the RNA genome and forms the internal core which is surrounded by the envelope in the virion. The C protein structure contains four alpha helices and forms dimers that are organized into tetramers. The tetramers form extended filamentous ribbons resembling the stacked alpha helices seen in HEAT protein structures.
Organizational Affiliation:
Institute of Molecular and Cell Biology, Singapore, Republic of Singapore. dokland@uab.edu