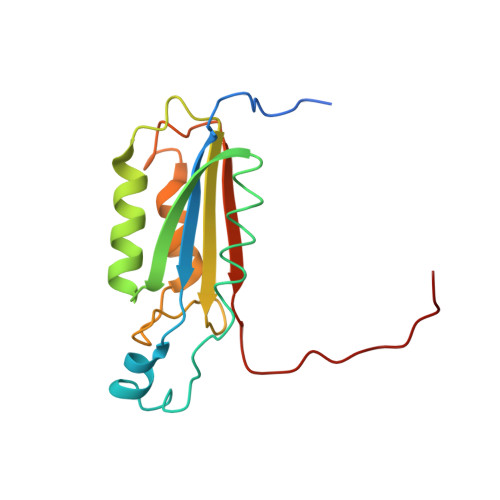
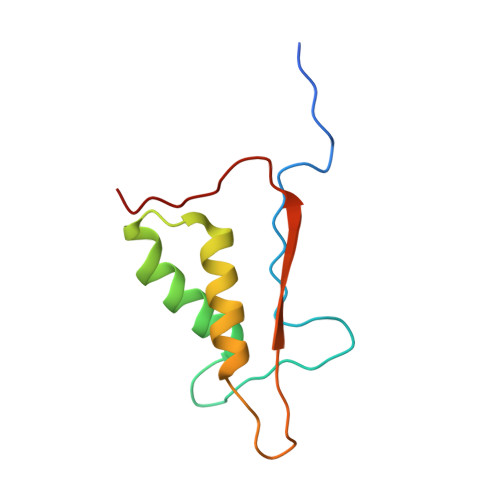
The atomic-resolution structure of human caspase-8, a key activator of apoptosis.
Watt, W., Koeplinger, K.A., Mildner, A.M., Heinrikson, R.L., Tomasselli, A.G., Watenpaugh, K.D.(1999) Structure 7: 1135-1143
- PubMed: 10508785
- DOI: https://doi.org/10.1016/s0969-2126(99)80180-4
- Primary Citation of Related Structures:
1QTN - PubMed Abstract:
Caspases are a family of cysteine proteases that have important intracellular roles in inflammation and apoptosis. Caspase-8 activates downstream caspases which are unable to carry out autocatalytic processing and activation. Caspase-8 is designated as an initiator caspase and is believed to sit at the apex of the Fas- or TNF-mediated apoptotic cascade. In view of this role, the enzyme is an attractive target for the design of inhibitors aimed at blocking the undesirable cell death associated with a range of degenerative disorders.
Organizational Affiliation:
Structural, Analytical & Medicinal Chemistry Pharmacia & Upjohn Inc. 301 Henrietta Street, Kalamazoo, MI 49007, USA.