The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 A resolution
Dyer, D.H., Rubio, L.M., Thoden, J.B., Holden, H.M., Ludden, P.W., Rayment, I.(2003) J Biol Chem 278: 32150-32156
- PubMed: 12754195
- DOI: https://doi.org/10.1074/jbc.M304264200
- Primary Citation of Related Structures:
1P90 - PubMed Abstract:
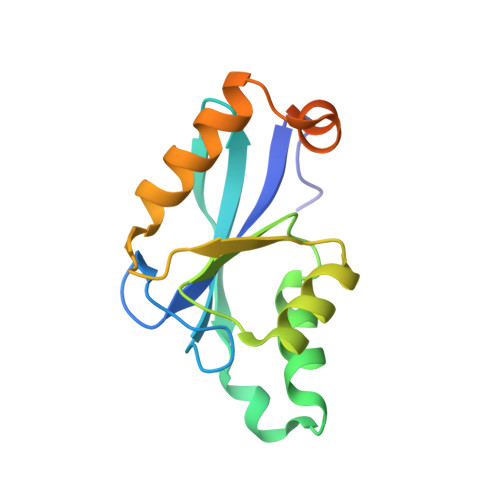
The Azotobacter vinelandii NafY protein (nitrogenase accessory factor Y) is able to bind either to the iron molybdenum cofactor (FeMo-co) or to apodinitrogenase and is believed to facilitate the transfer of FeMo-co into apodinitrogenase. The NafY protein has two domains: an N-terminal domain (residues Met1-Leu98) and a C-terminal domain (residues Glu99-Ser232), referred here to as the "core domain." The core domain of NafY is shown here to be capable of binding the FeMo cofactor of nitrogenase but unable to bind to apodinitrogenase in the absence of the first domain. The three-dimensional molecular structure of the core domain of NafY has been solved to 1.8-A resolution, revealing that the protein consists of a mixed five-stranded beta-sheet flanked by five alpha-helices that belongs to the ribonuclease H superfamily. As such, this represents a new fold capable of binding FeMo-co, where the only previous example was that seen in dinitrogenase.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison, Wisconsin 53706, USA.